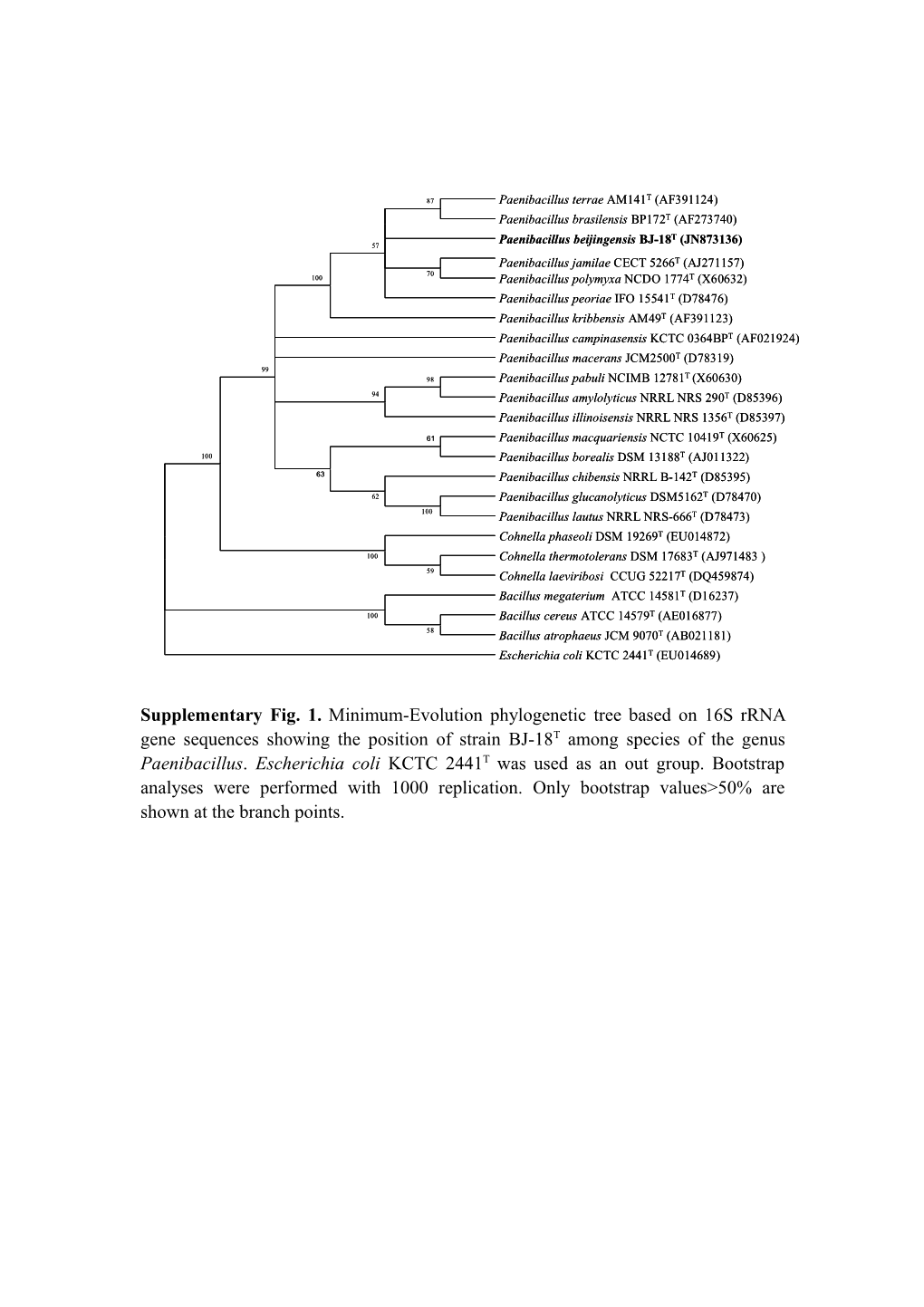
87 Paenibacillus terrae AM141T (AF391124) Paenibacillus brasilensis BP172T (AF273740) T 57 Paenibacillus beijingensis BJ-18 (JN873136) Paenibacillus jamilae CECT 5266T (AJ271157) 70 100 Paenibacillus polymyxa NCDO 1774T (X60632) Paenibacillus peoriae IFO 15541T (D78476) Paenibacillus kribbensis AM49T (AF391123) Paenibacillus campinasensis KCTC 0364BPT (AF021924) Paenibacillus macerans JCM2500T (D78319) 99 T 98 Paenibacillus pabuli NCIMB 12781 (X60630) 94 Paenibacillus amylolyticus NRRL NRS 290T (D85396) Paenibacillus illinoisensis NRRL NRS 1356T (D85397)
61 Paenibacillus macquariensis NCTC 10419T (X60625) 100 Paenibacillus borealis DSM 13188T (AJ011322) 63 Paenibacillus chibensis NRRL B-142T (D85395) 62 Paenibacillus glucanolyticus DSM5162T (D78470) 100 Paenibacillus lautus NRRL NRS-666T (D78473) Cohnella phaseoli DSM 19269T (EU014872) 100 Cohnella thermotolerans DSM 17683T (AJ971483 ) 59 Cohnella laeviribosi CCUG 52217T (DQ459874) Bacillus megaterium ATCC 14581T (D16237) 100 Bacillus cereus ATCC 14579T (AE016877) 58 Bacillus atrophaeus JCM 9070T (AB021181) Escherichia coli KCTC 2441T (EU014689)
Supplementary Fig. 1. Minimum-Evolution phylogenetic tree based on 16S rRNA gene sequences showing the position of strain BJ-18T among species of the genus Paenibacillus. Escherichia coli KCTC 2441T was used as an out group. Bootstrap analyses were performed with 1000 replication. Only bootstrap values>50% are shown at the branch points. T 73 Paenibacillus jamilae CECT 5266 (AJ271157) Paenibacillus polymyxa NCDO 1774T (X60632) Paenibacillus peoriae IFO 15541T (D78476) 99 52 Paenibacillus kribbensis AM49T (AF391123) Paenibacillus terrae AM141T (AF391124) 81 Paenibacillus brasilensis BP172T (AF273740) Paenibacillus beijingensis BJ-18T (JN873136) Paenibacillus illinoisensis NRRL NRS 1356T (D85397) 96 Paenibacillus pabuli NCIMB 12781T (X60630) 78 93 Paenibacillus amylolyticus NRRL NRS 290T (D85396) Paenibacillus macerans JCM2500T (D78319) Paenibacillus campinasensis KCTC 0364BPT (AF021924) T 92 Paenibacillus glucanolyticus DSM5162 (D78470) T 96 Paenibacillus lautus NRRL NRS-666 (D78473) Paenibacillus chibensis NRRL B-142T (D85395) Paenibacillus macquariensis NCTC 10419T (X60625) 57 Paenibacillus borealis DSM 13188T (AJ011322) Cohnella laeviribosi CCUG 52217T (DQ459874) 99 Cohnella thermotolerans DSM 17683T (AJ971483 ) 66 Cohnella phaseoli DSM 19269T (EU014872) Bacillus atrophaeus JCM 9070T (AB021181) T 100 Bacillus cereus ATCC 14579 (AE016877) Bacillus megaterium ATCC 14581T (D16237) Escherichia coli KCTC 2441T (EU014689)
Supplementary Fig. 2. Maximum-Likelihood phylogenetic tree based on 16S rRNA gene sequences showing the position of strain BJ-18T among species of the genus Paenibacillus. Escherichia coli KCTC 2441T was used as an out group. Bootstrap analyses were performed with 1000 replication. Only bootstrap values>50% are shown at the branch points. DPG
PME PE PG
Supplementary Fig. 3. Polar lipids of strain BJ-18T analyzed by TLC. Diphosphatidylglycerol (DPG), phosphatidylmethylethanolamine (PME), phosphatidylethanolamine (PE), phosphatidylglycerol (PG). Paenibacillus catalpae DSM 24714T (HQ657320) 99
88 Paenibacillus glycanilyticus JCM 11221T (AB042938)
T 55 Paenibacillus xinjiangensis DSM 16970 (AY839868)
Paenibacillus castaneae DSM 19417T (EU099594) 98 54 Paenibacillus prosopidis DSM 22405T (FJ820995)
Paenibacillus algorifonticola JCM 16598T (GQ383922)
Paenibacillus sputi DSM 22699T (FN394513)
Paenibacillus harenae DSM 16969T (AY839867)
92 Paenibacillus alkaliterrae DSM 17040T (AY960748)
78 Paenibacillus agarexedens DSM 1327T (AJ345020)
Paenibacillus chungangensis CCUG 59129T (GU187432)
Paenibacillus thailandensis KCTC 13043T (AB265205)
T 91 Paenibacillus agaridevorans DSM 1355 (AJ345023) 54 Paenibacillus nanensis KCTC 13044T (AB265206)
Paenibacillus granivorans A30T (AF237682)
Paenibacillus camelliae KCTC 13220T (EU400621) 99
T 74 Paenibacillus septentrionalis KCTC 13039 (AB295647)
52 Paenibacillus montaniterrae KCTC 13036T (AB295646)
96 Paenibacillus siamensis KCTC 13038T (AB295645)
Paenibacillus tarimensis DSM 19409T (EF125184)
Paenibacillus pasadenensis ATCC BAA-1211T (AY167820) 82
93 Paenibacillus humicus DSM 18784T (AM411528)
65 Paenibacillus wooponensis JCM 16350T (EU939687)
61 58 Paenibacillus beijingensis CGMCC 1.12048T (JN408293)
Paenibacillus tarimensis DSM 19409T (EF125184)
56 Paenibacillus pinihumi JCM 16419T (GQ423057)
Paenibacillus phyllosphaerae PALXIL04T (AY598818) 58
Paenibacillus sacheonensis DSM 23054T (GU124597)
Paenibacillus marinum BCRC 17723T (FR865169)
Paenibacillus sepulcri CCM 7311T (DQ291142) 94
Paenibacillus mendelii CCM 4839T (AF537343)
Paenibacillus sacheonensis DSM 23054T (GU124597)
99 Paenibacillus xylaniclasticus BCRCT (FJ532373)
Paenibacillus cellulosilyticus LMG 22232T (DQ407282)
89 Paenibacillus kobensis DSM 10249T (AB073363)
Paenibacillus thiaminolyticus NBRC 15656T (AB073197) 85
73 Paenibacillus dendritiformis CIP 105967T (AY359885)
99 Paenibacillus popilliae ATCC 14706T (AF071859)
Paenibacillus lentimorbus ATCC 14707T (AF071861)
Paenibacillus taiwanensis BCRC 17411T (DQ890521) 84 73 Paenibacillus assamensis J CM 13186T (AY884046)
Paenibacillus profundus BCRC 17543T (AB712351) 66 99
Paenibacillus apiarius NRRL NRS-1438T (U49247)
Paenibacillus tyraminigenes BCRC 17765T (AY728023)
95 Paenibacillus alvei DSM 29T (AMBZ01000001)
99 Paenibacillus alvei DSM 29T (AJ320491)
T 99 Paenibacillus senegalensis JCM66 (JF824808)
Paenibacillus residui DSM 22072T (FN293173)
Paenibacillus contaminans BCRC 17728T (EF626690)
T 99 Paenibacillus pueri KCTC 13223 (EU391156)
Paenibacillus thermoaerophilus JCMT (AB738879)
Paenibacillus thermoaerophilus BCRC 177842T (AB738878)
Paenibacillus larvae DSM 7030T (AY530294)
Paenibacillus ginsengarvi DSM 18677T (AB271057) 99
65 Paenibacillus hodogayensis JCM 12520T (AB179866)
Paenibacillus koleovorans JCM 11186T (AB041720)
T 66 Paenibacillus pocheonensis KCTC 13941 (AB245386)
Paenibacillus chondroitinus DSM 5051T (D82064) 88
Paenibacillus frigoriresistens JCM 18141T (JQ314346) 96 99 Paenibacillus alginolyticus DSM5050T (D78465) 99 Paenibacillus pectinilyticus KCTC 13222T (EU391157) 51 Paenibacillus aestuarii JCM 15521T (EU570250)
Paenibacillus chartarius CCUG 55240T (FN689718)
Paenibacillus chitinolyticus IFO 15660T (AB021183)
Paenibacillus gansuensis DSM 16968T (AY839866)
T 98 Paenibacillus ehimensis KCTC 3748 (AY116665)
Paenibacillus koreensis KCCM 40903T (AF130254)
Paenibacillus tianmuensis DSM 22342T (FJ719490)
Paenibacillus tianmuensis DSM 22342T (FJ719492) 88 97 Paenibacillus tianmuensis DSM 22342T (FJ719491)
Paenibacillus elgii NBRC 100335T (AY090110)
T 95 Paenibacillus rigui KCTC 13282 (EU939688)
Paenibacillus vulneris CCUG 53270T (HE649498)
Paenibacillus chinjuensis JCM 10939T (AF164345)
Paenibacillus mucilaginosus VKPM B-7519T (FJ039528) 99
62 Paenibacillus mucilaginosus KCTC 3870 T (AF006077)
Paenibacillus edaphicus DSM 12974 T (AF006076)
Paenibacillus soli LMG 23604T (DQ309072)
Paenibacillus filicis JCM 16417T (GQ423055)
Paenibacillus validus JCM 9077T (AB073203)
77 Paenibacillus naphthalenovorans DSM 14203T (AF353681)
64 Paenibacillus xylanisolvens KCTC 13042T (AB495094)
Paenibacillus sediminis DSM 23491T (GQ355277)
Paenibacillus terrigena JCM 21741T (AB248087)
Paenibacillus anaericanus DSM 15890T (AJ318909) 67
Paenibacillus pini JCM 16418T (GQ423056)
Paenibacillus turicensis DSM 14349T (AF378694)
T 53 Paenibacillus barengoltzii ATCC BAA-1209 (AY167814)
84 Paenibacillus phoenicis NBRC 106274T (EU977789)
T 87 Paenibacillus timonensis CCUG 48216 (AY323610)
Paenibacillus macerans IAM 12467T (AB073196)
99 Paenibacillus thermophilus DSM 24746T (JQ824133)
Paenibacillus konsidensis JCM 14798T (EU081509)
Paenibacillus puldeungensis KCTC 13718T (GU187433)
56 Paenibacillus sanguinis CIP 107938T (AY323609)
Paenibacillus woosongensis DSM 16971T (AY847463)
76 Paenibacillus fonticola LMG 23577T (DQ453131)
Paenibacillus telluris CGMCC 1.10695T (HQ257247)
Paenibacillus motobuensis CCUG 50090T (AY741810)
Paenibacillus chibensis JCM 9905T (AB073194) 99
Paenibacillus chibensis NRRL B-142T (D85395)
Paenibacillus cookii LMG 18419T (AJ250317)
82 Paenibacillus azoreducens DSM 13822T (AJ272249)
Paenibacillus cellulositrophicus KCTC 13135T (FJ178001) 67
T 99 Paenibacillus favisporus LMG 20987 (AY208751)
99 Paenibacillus cineris LMG 18439T (AJ575658)
Paenibacillus rhizosphaerae CECAP06T (AY751754)
Paenibacillus borealis CCUG 43137T (AJ011322)
Paenibacillus odorifer DSM 15391T (AJ223990)
Paenibacillus typhae DSM 25190T (JN256679)
52 53 Paenibacillus taohuashanense DSM 24089T (JQ694712)
Paenibacillus wynnii LMG 22176T (AJ633647) 60 87 Paenibacillus donghaensis JCM 86791T (EF079062)
Paenibacillus jilunlii DSM 23019T (GQ985393)
T 99 Paenibacillus graminis DSM 15220 (AJ223987)
95 Paenibacillus sonchi CCBAU 83901T (DQ358736)
84 Paenibacillus riograndensis CECT 7330T (EU257201)
Paenibacillus stellifer DSM 14472T (AJ316013)
T 87 Paenibacillus durus DSM 1735 (X77846)
51 83 Paenibacillus sophorae CGMCC 1.10238 T (GQ985395)
Paenibacillus zanthoxyli DSM 18202T (DQ471303)
97 Paenibacillus forsythiae DSM 17842T (DQ338443)
85 Paenibacillus sabinae DSM 17841T (DQ338444)
86 Paenibacillus brassicae JCM26112T (JN408294)
T 99 Paenibacillus uliginis LMG 24790 (DQ471303)
65 Paenibacillus purispatii DSM 22991T (EU888513)
Paenibacillus campinasensis JCM 11200T (AF021924) 88
Paenibacillus lactis DSM 15596T (AY257868)
92 Paenibacillus lautus NRRL NRS-666T (D78473)
99 Paenibacillus glucanolyticus DSM 5162T (AB073189)
T 60 Paenibacillus jamilae CECT 5266 (AJ271157)
54 Paenibacillus polymyxa ATCC 842T (AFOX01000032)
66 Paenibacillus peoriae DSM 8320T (AJ320494)
63 Paenibacillus brasilensis DSM 14914T (AF273740)
65 Paenibacillus beijingnica BJ-18T (JN408292)
99 Paenibacillus kribbensis JCM 11465T (AF391123)
66 Paenibacillus terrae JCM 11466T (AF391124)
61 Paenibacillus hunanensis CGMCC 1.8907T (EU741036)
Paenibacillus hordei KACC 15511T (HQ833590)
T 98 Paenibacillus urinalis CCUG 53521 (EF212892)
Paenibacillus provencensis CCUG 53519T (EF212893)
Paenibacillus massiliensis CCUG 48215T (AY230766)
99 Paenibacillus panacisoli KCTC 13020T (AB245384)
Paenibacillus illinoisensis NRRL NRS-1356T (D85397) 93
Paenibacillus xylanilyticus CECT 5839T (AY427832)
T 99 Paenibacillus pabuli JCM 9074 (AB073191)
Paenibacillus taichungensis DSM 19942T (EU179327) 91 51
Paenibacillus barcinonensis DSM 15478T (AJ716019)
99 Paenibacillus oceanisediminis JCM 17814T (JF811909)
Paenibacillus xylanexedens DSM 21292T (EU558281) 99
Paenibacillus amylolyticus NRRL NRS-290T (D85396)
99 Paenibacillus tundrae DSM 21291T (EU558284)
99 Paenibacillus tylopili BCRC 177241T (EF206295)
Paenibacillus nematophilus DSM 13559T (AF480935)
Paenibacillus glacialis DSM 22343T (EU815300)
T 99 Paenibacillus antarcticus LMG 22078 (AJ605292)
99 Paenibacillus macquariensis subsp. defensor JCM 14954T (AB360546)
97 Paenibacillus macquariensis subsp. macquariensis CCUG 37394T (X60625)
Bacillus subtilis NCDO 1769T (X60646)
0.01 Supplementary Fig. 4. Neighbor-joining phylogenetic tree based on 16Sr RNA gene sequences showing the position of strain BJ-18T (marked in red) among species of the genus Paenibacillus. Bacillus subtilis NCDO 1769T (X60646) was used as an out group. Bootstrap analyses were performed with 1000 cycles. Only bootstrap values >50% are shown at the branch points. Bar 0.01 substitutions per nucleotide positions.