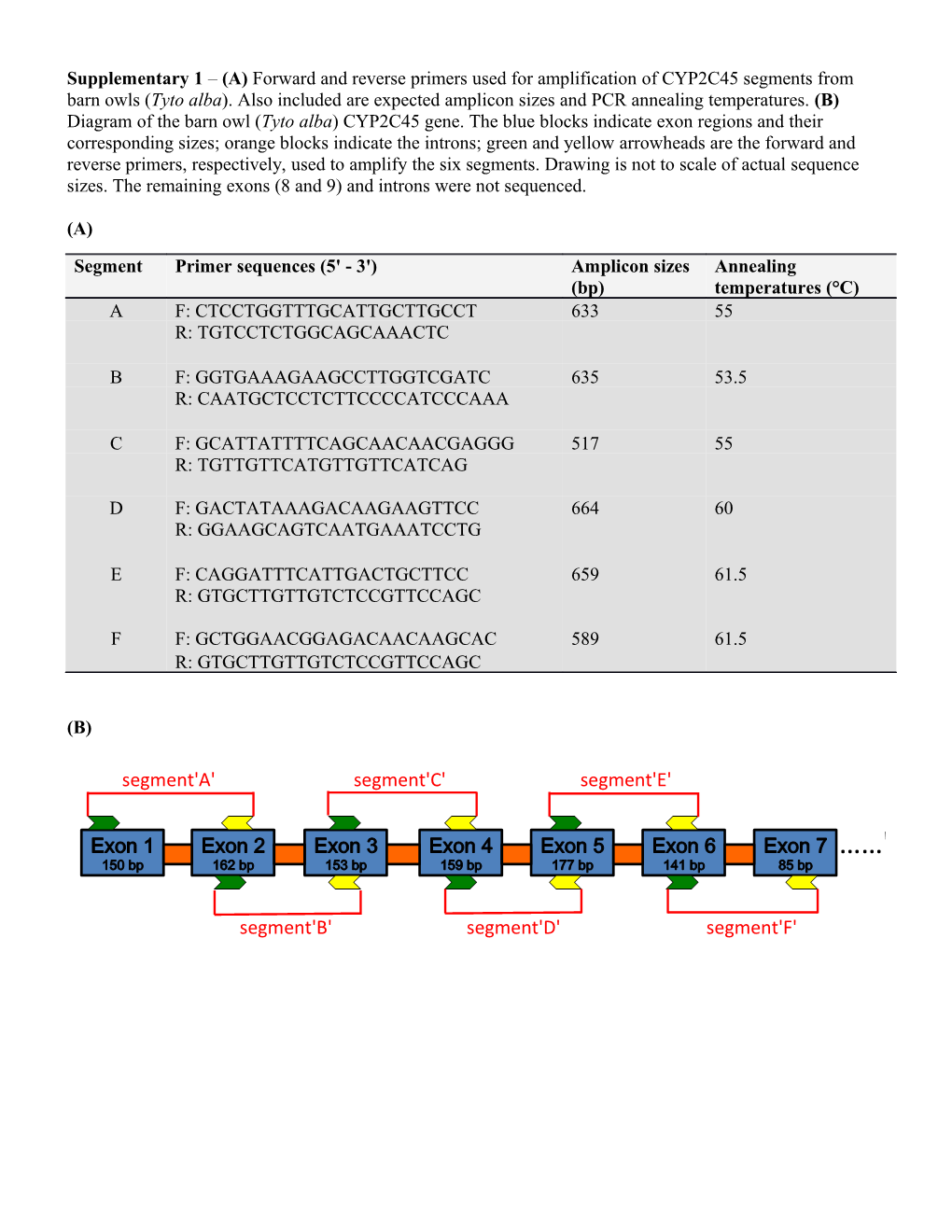
Supplementary 1 – (A) Forward and reverse primers used for amplification of CYP2C45 segments from barn owls (Tyto alba). Also included are expected amplicon sizes and PCR annealing temperatures. (B) Diagram of the barn owl (Tyto alba) CYP2C45 gene. The blue blocks indicate exon regions and their corresponding sizes; orange blocks indicate the introns; green and yellow arrowheads are the forward and reverse primers, respectively, used to amplify the six segments. Drawing is not to scale of actual sequence sizes. The remaining exons (8 and 9) and introns were not sequenced.
(A)
Segment Primer sequences (5' - 3') Amplicon sizes Annealing (bp) temperatures (°C) A F: CTCCTGGTTTGCATTGCTTGCCT 633 55 R: TGTCCTCTGGCAGCAAACTC
B F: GGTGAAAGAAGCCTTGGTCGATC 635 53.5 R: CAATGCTCCTCTTCCCCATCCCAAA
C F: GCATTATTTTCAGCAACAACGAGGG 517 55 R: TGTTGTTCATGTTGTTCATCAG
D F: GACTATAAAGACAAGAAGTTCC 664 60 R: GGAAGCAGTCAATGAAATCCTG
E F: CAGGATTTCATTGACTGCTTCC 659 61.5 R: GTGCTTGTTGTCTCCGTTCCAGC
F F: GCTGGAACGGAGACAACAAGCAC 589 61.5 R: GTGCTTGTTGTCTCCGTTCCAGC
(B)
segment'A' segment'C' segment'E'
……'
segment'B' segment'D' segment'F' Supplementary 2 – Summary of AICC statistics for models examining the relationship between various predictors and (a) total SGAR residue concentration, (b) presence/absence of any SGAR, (c) presence/absence of bromadiolone, and (d) presence/absence of toxicosis symptoms.
AICc Model AICC Δ AICC df weight (a) Total SGAR residue Concentration demographic + time.period -78.2 0 5 0.86 demographic*time.period -74.6 3.7 7 0.14
(b) Presence/absence of any SGAR demographic + time.period 154.7 0 5 0.82 demographic*time.period 157.7 3 7 0.18
(c) Presence/absence of bromadiolone demographic + time.period 153.6 0 5 0.85 demographic*time.period 157.1 3.5 7 0.15
(d) Presence/absence of toxicosis symptoms [bromadiolone] + [brodifacoum]*[difethialone]*demographic + time.period 42.5 0.0 15.0 0.9945 [bromadiolone]*[difethialone]*time.period + [brodifacoum] + demographic 55.5 13.0 12.0 0.0015 [bromadiolone]*time.period + [brodifacoum] + [difethialone] + demographic 56.7 14.1 12.0 <0.001 [bromadiolone] + [brodifacoum]*[difethialone]*time.period + demographic 57 14.4 9.0 <0.001 [bromadiolone] + [brodifacoum] + [difethialone]*time.period + demographic 57.5 15.0 9.0 <0.001 [bromadiolone]*[difethialone] + [brodifacoum] + demographic + time.period 58.5 16.0 9.0 <0.001 [bromadiolone] + [brodifacoum] + [difethialone]*demographic + time.period 58.6 16.1 10.0 <0.001 [bromadiolone] + [brodifacoum]*demographic + [difethialone] + time.period 59.1 16.6 10.0 <0.001 [bromadiolone] + [brodifacoum] + [difethialone] + demographic + time.period 59.4 16.8 9.0 <0.001 [bromadiolone]*[brodifacoum]*demographic + [difethialone] + time.period 59.4 16.9 8.0 <0.001 [bromadiolone] + [brodifacoum]*time.period + [difethialone] + demographic 60 17.5 12.0 <0.001 [bromadiolone]*demographic*time.period + [brodifacoum] + [difethialone] 61.2 18.6 9.0 <0.001 [bromadiolone]*[brodifacoum] + [difethialone] + demographic + time.period 61.6 19.0 9.0 <0.001 [bromadiolone]*[brodifacoum]*time.period + [difethialone] + demographic 61.7 19.1 10.0 <0.001 [bromadiolone]*demographic + [brodifacoum] + [difethialone] + time.period 62 19.5 15.0 <0.001 [bromadiolone] + [brodifacoum]*[difethialone] + time.period + demographic 62.5 20.0 14.0 <0.001 [bromadiolone] + [brodifacoum] + [difethialone] + demographic*time.period 62.8 20.2 10.0 <0.001 [bromadiolone]*[brodifacoum]*[difethialone] + demographic + time.period 64.9 22.4 12.0 <0.001 [bromadiolone]*[difethialone]*demographic + [brodifacoum] + time.period 65.9 23.4 15 <0.001 [bromadiolone] + [brodifacoum] + [difethialone]*demographic*time.period 66.4 23.9 40 <0.001 [bromadiolone]*[brodifacoum]*[difethialone]*time.period*demographic 67 24.4 15 <0.001 Supplementary 3 – SNPs identified in the non-coding regions (introns) of the CYP2C45 gene and their associated nucleotide positions (starting from the start codon of exon 1). Highlighted in red are individuals categorized as “low AR tolerance”, and blue are “high AR tolerance”.
Nucleotide position 459 510 1023 1875 1913 1979 2662 3188 3223
L12-558 R Y R G C Y K Y G L12-559 R Y G G C T K Y R L12-709 R T G G C T K C G L12-736 A C A G C T G T G L12-753 G T G G C T T C R L12-727 G T G R A T K C G L12-737 A Y R G C T G T G L12-738 R T G G C T K C R L12-555 R Y G G C T K Y R L12-732 G T G G A T K C G L12-733 R C A G C T G Y G L12-752 G T G R C Y T C G L12-949 R Y R R C T K Y G