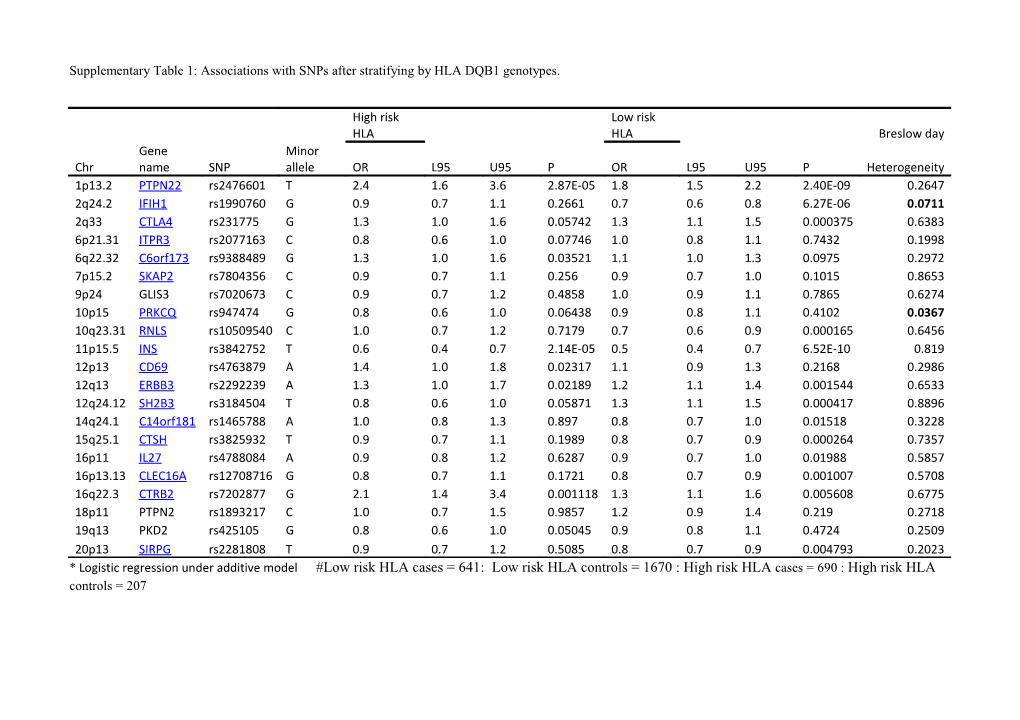
Supplementary Table 1: Associations with SNPs after stratifying by HLA DQB1 genotypes.
High risk Low risk HLA HLA Breslow day Gene Minor Chr name SNP allele OR L95 U95 P OR L95 U95 P Heterogeneity 1p13.2 PTPN22 rs2476601 T 2.4 1.6 3.6 2.87E-05 1.8 1.5 2.2 2.40E-09 0.2647 2q24.2 IFIH1 rs1990760 G 0.9 0.7 1.1 0.2661 0.7 0.6 0.8 6.27E-06 0.0711 2q33 CTLA4 rs231775 G 1.3 1.0 1.6 0.05742 1.3 1.1 1.5 0.000375 0.6383 6p21.31 ITPR3 rs2077163 C 0.8 0.6 1.0 0.07746 1.0 0.8 1.1 0.7432 0.1998 6q22.32 C6orf173 rs9388489 G 1.3 1.0 1.6 0.03521 1.1 1.0 1.3 0.0975 0.2972 7p15.2 SKAP2 rs7804356 C 0.9 0.7 1.1 0.256 0.9 0.7 1.0 0.1015 0.8653 9p24 GLIS3 rs7020673 C 0.9 0.7 1.2 0.4858 1.0 0.9 1.1 0.7865 0.6274 10p15 PRKCQ rs947474 G 0.8 0.6 1.0 0.06438 0.9 0.8 1.1 0.4102 0.0367 10q23.31 RNLS rs10509540 C 1.0 0.7 1.2 0.7179 0.7 0.6 0.9 0.000165 0.6456 11p15.5 INS rs3842752 T 0.6 0.4 0.7 2.14E-05 0.5 0.4 0.7 6.52E-10 0.819 12p13 CD69 rs4763879 A 1.4 1.0 1.8 0.02317 1.1 0.9 1.3 0.2168 0.2986 12q13 ERBB3 rs2292239 A 1.3 1.0 1.7 0.02189 1.2 1.1 1.4 0.001544 0.6533 12q24.12 SH2B3 rs3184504 T 0.8 0.6 1.0 0.05871 1.3 1.1 1.5 0.000417 0.8896 14q24.1 C14orf181 rs1465788 A 1.0 0.8 1.3 0.897 0.8 0.7 1.0 0.01518 0.3228 15q25.1 CTSH rs3825932 T 0.9 0.7 1.1 0.1989 0.8 0.7 0.9 0.000264 0.7357 16p11 IL27 rs4788084 A 0.9 0.8 1.2 0.6287 0.9 0.7 1.0 0.01988 0.5857 16p13.13 CLEC16A rs12708716 G 0.8 0.7 1.1 0.1721 0.8 0.7 0.9 0.001007 0.5708 16q22.3 CTRB2 rs7202877 G 2.1 1.4 3.4 0.001118 1.3 1.1 1.6 0.005608 0.6775 18p11 PTPN2 rs1893217 C 1.0 0.7 1.5 0.9857 1.2 0.9 1.4 0.219 0.2718 19q13 PKD2 rs425105 G 0.8 0.6 1.0 0.05045 0.9 0.8 1.1 0.4724 0.2509 20p13 SIRPG rs2281808 T 0.9 0.7 1.2 0.5085 0.8 0.7 0.9 0.004793 0.2023 * Logistic regression under additive model #Low risk HLA cases = 641: Low risk HLA controls = 1670 : High risk HLA cases = 690 : High risk HLA controls = 207