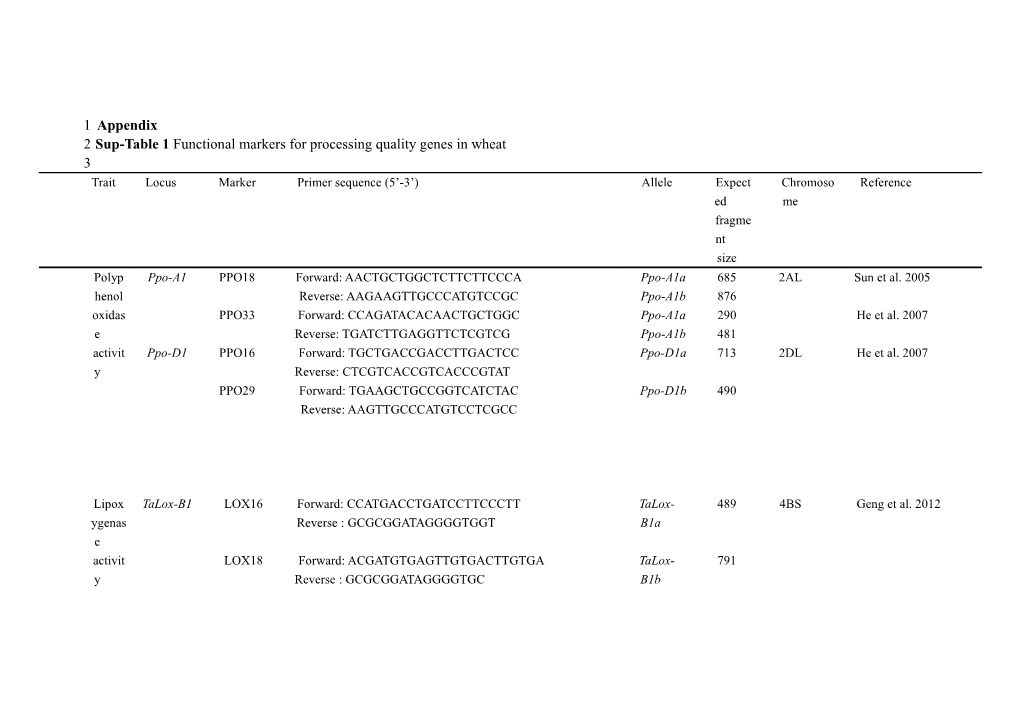
1 Appendix 2 Sup-Table 1 Functional markers for processing quality genes in wheat 3 Trait Locus Marker Primer sequence (5’-3’) Allele Expect Chromoso Reference ed me fragme nt size Polyp Ppo-A1 PPO18 Forward: AACTGCTGGCTCTTCTTCCCA Ppo-A1a 685 2AL Sun et al. 2005 henol Reverse: AAGAAGTTGCCCATGTCCGC Ppo-A1b 876 oxidas PPO33 Forward: CCAGATACACAACTGCTGGC Ppo-A1a 290 He et al. 2007 e Reverse: TGATCTTGAGGTTCTCGTCG Ppo-A1b 481 activit Ppo-D1 PPO16 Forward: TGCTGACCGACCTTGACTCC Ppo-D1a 713 2DL He et al. 2007 y Reverse: CTCGTCACCGTCACCCGTAT PPO29 Forward: TGAAGCTGCCGGTCATCTAC Ppo-D1b 490 Reverse: AAGTTGCCCATGTCCTCGCC
Lipox TaLox-B1 LOX16 Forward: CCATGACCTGATCCTTCCCTT TaLox- 489 4BS Geng et al. 2012 ygenas Reverse : GCGCGGATAGGGGTGGT B1a e activit LOX18 Forward: ACGATGTGAGTTGTGACTTGTGA TaLox- 791 y Reverse : GCGCGGATAGGGGTGC B1b Yello Psy-A1 YP7A Forward: GGACCTTGCTGATGACCGAG PsyA1a 194 7AL He et al. 2008 w Reverse: TGACGGTCTGAAGTGAGAATGA PsyA1b 231 pigme YP7A-2 Forward: GCCAGCCCTTCAAGGACATG PsyA1a 1686 He et al. 2009a nt Reverse: CAGATGTCGCCACACTGCCA PsyA1b 1686 conten PsyA1c 1001 t Psy-B1 YP7B-1 Forward: GCCACAACTTGAATGTGAAAC Psy-B1a 151 7BL He et al. 2009a Reverse: ACTTCTTCCATTTGAACCCC Psy-B1b 156 YP7B-2 Forward: GCCACCCACTGATTACCACTA Psy-B1c 428 Reverse: CCAAGGTGAGGGTCTTCAAC YP7B-3 Forward: GAGTAAGCCACCCACTGATT Psy-B1d 884 Reverse: TCGCTGAGGAATGTACTGAC YP7B-4 Forward: AGGTACCAGCCAGCCCATA Psy-B1e 716 Reverse: CTCGTCAAAGTTCGTGTACC Psy1-D1 YP7D-1 Forward: TCCGACACCATCACCAAGTTCC Psy1-D1a 1074 7DL Wang JW et al. 2009a Reverse: CGTTGTAGGTTTGTGGGAGT Psy1-D1g 1093 YP7D-2 Forward: ACTCCCACAAACCTACAACG Psy1-D1a 967 Reverse: ACGCTCATCAACCCCACG Psy1-D1g 1046 TaZds-A1 YP2A-1 Forward: CCCTAAGGAAGCCGAGCAAAT TaZds- 183 2AL Dong et al. 2012 Reverse: GTGAGAGTACTAATGTTATGACCG A1a 179 TaZds- A1b TaZds-D1 YP2D-1 Forward: GTGGGATCCTGTTGCTTATGC TaZds- 2DL Zhang et al. 2011 Reverse: GTAGATTATCCAAGCCAACTGCC D1a 981 TaZds- D1b Bread Glu-A1 UMN19 UMN19F: CGAGACAATATGAGCAGCAAG Ax2* 344 1AL Liu SX et al. 2008b and UMN19R: CTGCCATGGAGAAGTTGGA Ax1 362 noodle Ax-null 362 - Ax2* Forward: ATGACTAAGCGGTTGGTTCTT Ax2* 1319 Ma et al. 2003 makin Reverse: ACCTTGCTCCCCTTGTCTTT g Glu-B1 bx7-f/r bx7-f: CACTGAGATGGCTAAGCGCC Bx-6 321 1BL Schwarz et al. 2004 quality bx7-r: GCCTTGGACGGCACCACAGG * TaBAC1215C06-F517: ACGTGTCCAAGCTTTGGTTC Bx7OE 447 Ragupathy et al. 2008 TaBAC1215C06-R964: GATTGGTGGGTGGATACAGG * TaBAC1215C06-F24671: CCACTTCCAAGGTGGGACTA Bx7OE 844 TaBAC1215C06-R25515: TGCCAACACAAAAGAAGCTG Bx Forward: CGCAACAGCCAGGACAATT nonBx17 630 Ma et al. 2003 Reverse: AGAGTTCTATCACTGCCTGGT Bx17 766 669 ZSBy8F5/R Forward: TTAGCGCTAAGTGCCGTCT By8 527 Lei et al. 2006 5 Reverse: TTGTCCTATTTGCTGCCCTT nonBy8
ZSBy9aF1/ Forward: TTCTCTGCATCAGTCAGGA By9 662 R3 Reverse: AGAGAAGCTGTGTAATGCC nonBy9 707
ZSBy9F7/R Forward: TACCCAGCTTCTCAGCAG By9 6 Reverse: TTGTCCCGACTGTTGTGG
ZSBy9F2/R Forward: GCAGTACCCAGCTTCTCAA By16 2 Reverse: CCTTGTCTTGTTTGTTGCC By null Bx20
Glu-D1 UMN25 UMN25F: GGGACAATACGAGCAGCAAA Dx2 299 1DL Liu SX et al. 2008b UMN25R: CTTGTTCCGGTTGTTGCCA Dx5 281 Dx5 Forward: CGTCCCTATAAAAGCCTAGC Dx5 478 Ma et al. 2003 Reverse: AGTATGAAACCTGCTGCGGAC * Forward: GCCTAGCAACCTTCACAATC Dx5 450 D΄Ovidio and Reverse: GAAACCTGCTGCGGACAAG Anderson 1994 UMN26 UMN26F: CGCAAGACAATATGAGCAAACT Dy10 397 Liu SX et al. 2008b UMN26R: TTGCCTTTGTCCTGTGTGC Dy12 415 P3 /P4 Forward: GTTGGCCGGTCGGCTGCCATG Dy10 576 Smith et al. 1994 Reverse: TGGAGAAGTTGGATAGTACC Dy12 612 5+10 Dx_F: TTTGGGGAATACCTGCACTACTAAAAAGGT Glu-D1d 343 Ishikawa and Dx5_F: AAAAGGTATTACCCAAGTGTAACTTGTCCG 320 Nakamura 2007 Dx_R: AATTGTCCTGGCTGCAGCTGCGA nonGlu- 361 D1d Glu-A3 gluA3a LA1F: AAACAGAATTATTAAAGCCGG Glu-A3a 529 1AS Wang LH et al. 2010 SA1R: GGTTGTTGTTGTTGCAGCA gluA3b LA3F: TTCAGATGCAGCCAAACAA Glu-A3b 894 SA2R: GCTGTGCTTGGATGATACTCTA gluA3d LA3F: TTCAGATGCAGCCAAACAA Glu-A3d 967 SA4R: TGGGGTTGGGAGACACATA gluA3e LA1F: AAACAGAATTATTAAAGCCGG Glu-A3e 158 SA5R: GGCACAGACGAGGAAGGTT gluA3f LA1F: AAACAGAATTATTAAAGCCGG Glu-A3f 552 SA6R: GCTGCTGCTGCTGTGTAAA gluA3g LA1F: AAACAGAATTATTAAAGCCGG Glu-A3g 1345 SA7R: AAACAACGGTGATCCAACTAA gluA3ac LA1F: AAACAGAATTATTAAAGCCGG Glu-A3a 573 SA3R: GTGGCTGTTGTGAAAACGA Glu-A3c Glu-B3 gluB3a SB1F: CACAAGCATCAAAACCAAGA Glu-B3a 1095 1BS Wang LH et al. 2009b SB1R: TGGCACACTAGTGGTGGTC gluB3b SB2F: ATCAGGTGTAAAAGTGATAG Glu-B3b 1570 SB2R: TGCTACATCGACATATCCA gluB3c SB3F: CAAATGTTGCAGCAGAGA Glu-B3c 472 SB3R: CATATCCATCGACTAAACAAA gluB3d SB4F: CACCATGAAGACCTTCCTCA Glu-B3d 662 SB4R: GTTGTTGCAGTAGAACTGGA gluB3e SB5F: GACCTTCCTCATCTTCGCA Glu-B3e 669 SB5R: GCAAGACTTTGTGGCATT gluB3g SB7F: CCAAGAAATACTAGTTAACACTAGTC Glu-B3g 853 SB7R: GTTGGGGTTGGGAAACA gluB3h SB8F: CCACCACAACAAACATTAA Glu-B3h 1022 SB8R: GTGGTGGTTCTATACAACGA gluB3i SB9F: TATAGCTAGTGCAACCTACCAT Glu-B3i 621 SB9R: TGGTTGTTGCGGTATAATTT gluB3bef SB10F: GCATCAACAACAAATAGTACTAGAA Glu-B3b 750 SB10R: GGCGGGTCACACATGACA Glu-B3e Glu-B3f gluB3fg SB6F: TATAGCTAGTGCAACCTACCAT Glu-B3f 812 SB6R: CAACTACTCTGCCACAACG Glu-B3g Grain Pin-a Pina-N2 Forward: TCAACATTCGTGCATCATCA Pina-D1r 436 5DS Chen et al. 2012 hardne Reverse: CTTCATTCGTCAGAGTTCCAT ss Pin-b * Forward: ATGAAGACCTTATTCCTCCTA Pinb-D1a 250 Giroux and Morris Reverse: CTCATGCTCACAGCCGCC 1997 * Forward: ATGAAGACCTTATTCCTCCTA Pinb-D1b 250 Reverse: CTCATGCTCACAGCCGCT
Starch Wx-B1 * Forward: CTGGCCTGCTACCTCAAGAGCAACT Wx-B1a 425 4AL Nakamura et al. 2002 proper Reverse: CTGACGTCCATGCCGTTGACGA ty Wild type BDFL: CTGGCCTGCTACCTCAAGAGCAACT Wx-B1 778 Saito et al. 2009 BRC1: GGTTGCGGTTGGGGTCGATGAC Null BFC: CGTAGTAAGGTGCAAAAAAGTGCCACG Null Wx- 668 BRC2: ACAGCCTTATTGTACCAAGACCCATGTGTG B1
4 * Unnamed marker 5 6 7 8 9 10 11 12 13Sup-Table 2 Functional markers for agronomic traits in wheat 14 Trait Locus Marker Primer sequence (5’-3’) Allele Expect Chromoso Reference ed me fragme nt size Reduction in Rht-B1 BF-WR1 BF: GGTAGGGAGGCGAGAGGCGAG Rht-B1a 237 4BS Ellis et al. 2002 plant height WR1: CATCCCCATGGCCATCTCGAGCTG BF-MR1 BF: GGTAGGGAGGCGAGAGGCGAG Rht-B1b 237 MR1: CATCCCCATGGCCATCTCGAGCTA Rht-D1 DF2- DF2: GGCAAGCAAAAGCTTCGCG Rht-D1a 264 4DS WR2 WR2: GGCCATCTCGAGCTGCAC
DF-MR2 DF: CGCGCAATTATTGGCCAGAGATAG Rht-D1b 254 MR2: CCCCATGGCCATCTCGAGCTGCTA Phototperiod Ppd-D1 * Ppd-D1_F: ACGCCTCCCACTACACTG Ppd-D1a 288 2DS Beales et al. response Ppd-D1_R2: CACTGGTGGTAGCTGAGATT 2007 Ppd-D1_R1: GTTGGTTCAAACAGAGAGC Ppd-D1b 414 Vernalization VRN-A1 * VRN1AF: GAAAGGAAAAATTCTGCTCG Vrn-A1a 965+87 5AL Yan et al. 2004a VRN1-INT1R: GCAGGAAATCGAAATCGAAG Vrn-A1b 6 Vrn-A1c 714 Vrn-A1 734 734
* Intr1/A/F2: AGCCTCCACGGTTTGAAAGTAA Vrn-A1c 1170 Fu et al. 2005 Intr1/A/R3: AAGTAAGACAACACGAATGTGAGA * Intr1/C/F: GCACTCCTAACCCACTAACC Vrn-A1 1068 Intr1/AB/R: TCATCCATCATCAAGGCAAA * Ex1/C/F: GTTCTCCACCGAGTCATGGT Vrn-A1 522 Intr1/A/R3: AAGTAAGACAACACGAATGTGAGA VRN-B1 * Intr1/B/F: CAAGTGGAACGGTTAGGACA Vrn-B1 709 5BL Intr1/B/R3: CTCATGCCAAAAATTGAAGATGA * Intr1/B/F: CAAGTGGAACGGTTAGGACA Vrn-B1 1149 Intr1/B/R4: CAAATGAAAAGGAATGAGAGCA * VRNBPF1: CCCCTGCTACCAGTGCCTACTA Vrn-B1 928 Chu et al. 2011 VBINSR1: CCCGCTTGTTGGCTGGTGAG * VRNBPF1: CCCCTGCTACCAGTGCCTACTA vrn-B1 989 VRNBPR1: GCCCCATCTCCGCTGGAGAACG VRN-D1 * Intr1/D/F: GTTGTCTGCCTCATCAAATCC Vrn-D1 1671 5DL Fu et al. 2005 Intr1/D/R3: GGTCACTGGTGGTCTGTGC * Intr1/D/F: GTTGTCTGCCTCATCAAATCC vrn-D1 997 Intr1/D/R4: AAATGAAAAGGAACGAGAGCG VRN-H1 * Intr1/H/F1: GCTCCAGCTGATGAAACTCC Vrn-H1 474 7HS Intr1/H/R1: CTTCATGGTTTTGCAAGCTCC * Intr1/H/F3: TTCATCATGGATCGCCAGTA Vrn-H1 403 Intr1/H/R3: AAAGCTCCTGCCAACTACGA VRN-B3 * VRN4-B-INS-F: CATAATGCCAAGCCGGTGAGTAC Vrn-B3 ~1200 7BS Yan et al. 2006 VRN4-B-INS-R: ATGTCTGCCAATTAGCTAGC * VRN4-BNOINS-F: ATGCTTTCGCTTGCCATCC Vrn-B3 ~1140 VRN4-BNOINS-R: CTATCCCTACCGGCCATTAG Kernel TaSus2- Sus2-SNP Sus2-SNP-185: TAAGCGATGAATTATGGC Hap-H 423 2BS Jiang et al. 2011 weight 2B - Sus2-SNP-589H2: GGTGTCCTTGAGCTTCTGG 185/589H 2 Sus2-SNP Sus2-SNP-227: CTATAGTATGAGCTGGATCAATGGC Hap-L 381 - Sus2-SNP-589L2: GGTGTCCTTGAGCTTCTGA 227/589L 2 TaGW2- Hap-6A- Forward: CGTTACCTCTGGTTTGGGTGTCGTG TaGW2- 949 6AS Su et al. 2011 6A P1 Reverse: CACCTCTCGAAAATCTTCCCAATTA 6A
Hap-6A- Forward: GAGAAAGGGCTGGTGCTATGGA TaGW2- 418 P2 Reverse: GTAACGCTTGATAAACATAGGTAAT 6A
TaCwi-A1 CWI22 Forward: GGTGATGAGTTCATGGTTAAT TaCwi- 402 2AL Ma et al. 2012 Reverse: AGAAGCCCAACATTAAATCAAC A1a
CWI21 Forward: GTGGTGATGAGTTCATGGTTAAG TaCwi- 404 Reverse: AGAAGCCCAACATTAAATCAAC A1b
Abiotic Dreb-B1 * P18F: CCCAACCCAAGTGATAATAATCT Dreb-B1 717 3BL Wei et al. 2009 stress P18R: TTGTGCTCCTCATGGGTACTT tolerance 15 16 * Unnamed marker 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 Sup-Table 3 Functional markers for disease resistance genes in wheat 33 Trait Locus Marke Primer sequence (5’-3’) Alle Expecte Chromosom Reference r le d e fragment size Powdery mildew Pm3 Pm3a Pm3a/F: GGAGTCTCTTCGCATAGA Pm 624 1AS Tommasini et al. resistance Pm3a/R: CAGCTTCTAAGATCAAGGAT 3a 2006
Pm3b Pm3b/F: GGCACAGACAAAGCTCTG Pm 1382 Pm3b/R: TCGAGTAGCTCGGGAATC 3b
Pm3c Pm3c/F: CTAGTGGAGGTAGTTGAC Pm 846 Pm3c/R: AGTCGTTCAAGAGAACGGC 3c
Pm3d Pm3d/F: TGACTATTCGTGGGTGCA Pm 1109 Pm3d/R: GACTGCGGCACAGTTCAGC 3d
Pm3e Pm3e/F: GGAATCCCTTTGGCTTGT Pm 524 Pm3e/R: CTAGCAGAGCAGTGCAAG 3e
Pm3f Pm3f/F: GGAGTCTCTTTGCTTAAG Pm 624 Pm3f/R: CAGCTTCTAAGATCAAGGAT 3f
Pm3g Pm3g/F: GAATCCCTTTATCTTGAC Pm 540 Pm3g/R: ATTCCCCTAGCAGAGCAGAA 3g
Leaf rust, Lr34/ cssfr1 L34DINT9F: TTGATGAAACCAGTTTTTTTTCTA +Lr 517 7DS Lagudah et al. 2009 stripe rust, and Yr18/ L34PLUSR: GCCATTTAACATAATCATGATGGA 34 powdery mildew Pm38 resistance cssfr2 L34DINT9F: TTGATGAAACCAGTTTTTTTTCTA - 523 L34MINUSR: TATGCCATTTAACATAATCATGAA Lr3 4 cssfr3 Lr34DINT9F: TTGATGAAACCAGTTTTTTTTCTA +Lr 517+150 Lr34PLUSR: GCCATTTAACATAATCATGATGGA 34 229 csLV34F: GTTGGTTAAGACTGGTGATGG - csLV34R: TGCTTGCTATTGCTGAATAGT Lr3 4
cssfr4 Lr34DINT9F: TTGATGAAACCAGTTTTTTTTCTA +Lr 150 Lr34MINUSR: TATGCCATTTAACATAATCATGAA 34 523+229 csLV34F: GTTGGTTAAGACTGGTGATGG - csLV34R: TGCTTGCTATTGCTGAATAGT Lr3 4
cssfr5 Lr34DINT9F: TTGATGAAACCAGTTTTTTTTCTA +Lr 751 Lr34MINUSR: TATGCCATTTAACATAATCATGAA 34 523 Lr34SPF: GGGAGCATTATTTTTTTCCATCATG - Lr34DINT13R2: ACTTTCCTGAAAATAATACAAGCA Lr3 4
cssfr6 cssfr6_f: CTGAGGCACTCTTTCCTGTACAAAG +Lr 652 cssfr6_r: GCATTCAATGAGCAATGGTTATC 34 649 - Lr3 4 cssfr7 cssfr7_f: GCGTATTGTAATGTATCGTGAGAG +Lr 247 cssfr7_r: CATAGGAATTTGTGTGCTGTCC 34 214 - Lr3 4 34 35 * Unnamed marker 36 37 38 39 40 41 42 43 44 45 46 47 Sup-Table 4 Functional markers for alien genes introduced in common wheat 48 Trait Locus Marker Primer sequence (5’-3’) Allele Expecte Chromosom Reference d e fragme nt size Bread- and 1BL·1RS ω-sec-p1 ω-sec-P1 (sense): ACCTTCCTCATCTTTGTCCT ω- 1100 1BL·1RS Froidmont 1998 noodle- /ω-sec-p2 ω-sec-P2 (antisense): CCGATGCCTATACCACTACT secalin making quality ω-sec-P3 ω-sec-P3 (sense): CCTTCCTCATCTTTGTCCTC ω- 400 /ω-sec-P4 ω-sec-P4 (antisense): secalin GCTCTGGTCTCTGGGGTTGT SECA2 Forward: GTTTGCTGGGGAATTATTTG 1BL·1 412 Chai et al. 2006 /SECA3 Reverse: TCCTCATCTTTGTCCTCGCC RS
H20 Forward: GTTGGAAGGGAGCTCGAGCTG 1BL·1 1598 Liu C et al. 2008a Reverse: GTTGGGCAGAAAGGTCGACATC RS
Grain protein Gpc- Xucw108 Forward: AGCCAGGGATAGAGGAGGAA Gpc- 217 6BS Uauy et al. 2006 content and B1 and Reverse: AGCTGTGAGCTGGTGTCCTT B1 and stripe rust Yr36 Xuhw89 UHW89-BF: TCTCCAAGAGGGGAGAGACA Yr36 126 Distelfeld et al. 2006 UHW89-R: TTCCTCTACCCATGAATCTAGCA Stripe rust Yr17 VENTRIUP/ VENTRIUP: AGGGGCTACTGACCA AGGCT 2NS 259 2NS·2AS Helguera et al. 2003 LN2 LN2: TGCAGCTACAGCAGTATGTACACA AAA URIC/LN2 URIC: GGTCGCCCTGGCTTGCACCT 2NS 285 LN2: TGCAGCTACAGCAGTATGTACACA AAA 2AS 275 Leaf rust Lr19 AG15F1/ AG15F1: AGCTCCTTGTGACTGAAATGAATG Lr19+ 184 7AgL·7AL Gennaro et al. 2009 AG15R1 AG15R1: AGATCTGCATTTTTAGCTTGACCA * AG15F4: CAGCTACGTGCATCCCTTTCTT AG15R1: AGATCTGCATTTTTAGCTTGACCA Lr19+ 811 AG15R5: GGAGGTACCTTTGCCCACTCA Lr19- 1073 Lr21 D14 Forward: CGCTTTTACCGAGATTGGTC Lr21 885 1DS Talbert et al. 1994 Reverse: CCAAAGAGCATCCATGGTGT Lr47 PS10R/ PS10L PS10R: GCTGATGACCCTGACCGGT Lr47 282 7S·7AS Helguera et al. 2000 PS10L: TCTTCATGCCCGGTCGGGT PS10R/ PS10R: GCTGATGACCCTGACCGGT 7S 450 PS10L2 PS10L2: GGGCAGGCGTTTATTCCAG Lr51 S30-13L/ S30-13L: GCATCAACAAGATATTCGTTATGACC 1S 819 1S·1BL Helguera et al. 2005 AGA7-759R AGA7-759R: TGGCTGCTCAGAAAACTGGACC 1B 783 49 50 * Unnamed marker 51 52 53 54