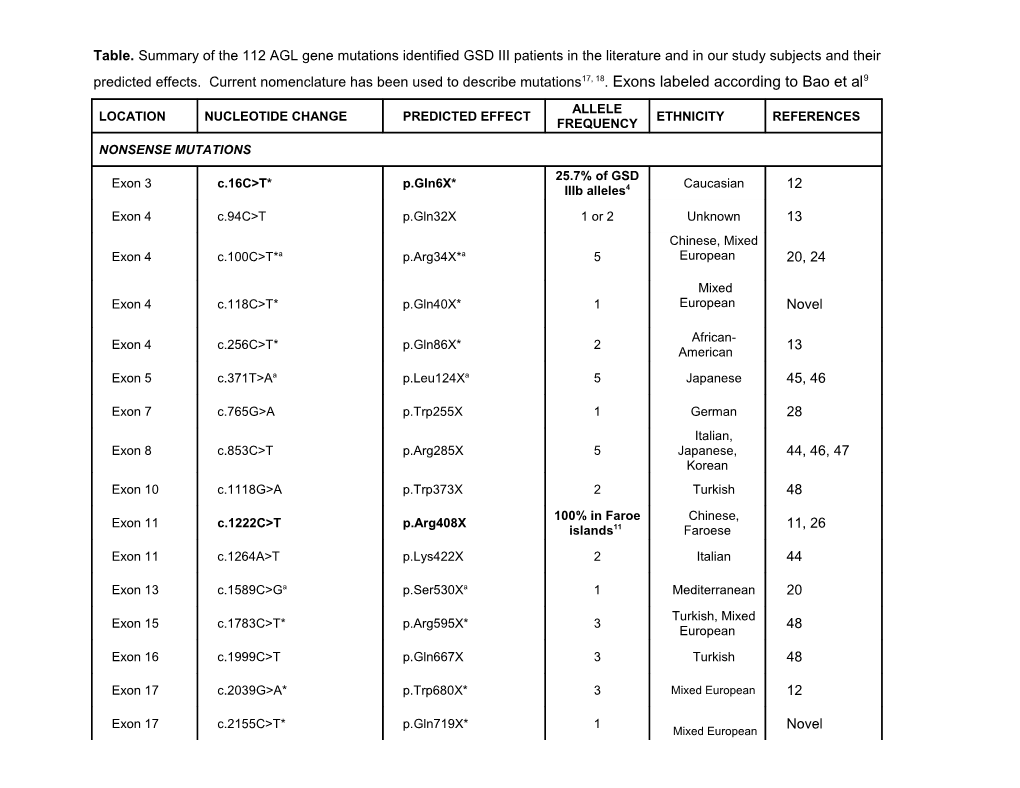
Table. Summary of the 112 AGL gene mutations identified GSD III patients in the literature and in our study subjects and their predicted effects. Current nomenclature has been used to describe mutations17, 18. Exons labeled according to Bao et al9
ALLELE LOCATION NUCLEOTIDE CHANGE PREDICTED EFFECT ETHNICITY REFERENCES FREQUENCY
NONSENSE MUTATIONS
25.7% of GSD Exon 3 c.16C>T* p.Gln6X* Caucasian 12 IIIb alleles4 Exon 4 c.94C>T p.Gln32X 1 or 2 Unknown 13 Chinese, Mixed Exon 4 c.100C>T*a p.Arg34X*a 5 European 20, 24
Mixed Exon 4 c.118C>T* p.Gln40X* 1 European Novel
African- Exon 4 c.256C>T* p.Gln86X* 2 13 American Exon 5 c.371T>Aa p.Leu124Xa 5 Japanese 45, 46
Exon 7 c.765G>A p.Trp255X 1 German 28 Italian, Exon 8 c.853C>T p.Arg285X 5 Japanese, 44, 46, 47 Korean Exon 10 c.1118G>A p.Trp373X 2 Turkish 48 100% in Faroe Chinese, Exon 11 c.1222C>T p.Arg408X 11, 26 islands11 Faroese Exon 11 c.1264A>T p.Lys422X 2 Italian 44
Exon 13 c.1589C>Ga p.Ser530Xa 1 Mediterranean 20 Turkish, Mixed Exon 15 c.1783C>T* p.Arg595X* 3 48 European Exon 16 c.1999C>T p.Gln667X 3 Turkish 48
Exon 17 c.2039G>A* p.Trp680X* 3 Mixed European 12
Exon 17 c.2155C>T* p.Gln719X* 1 Novel Mixed European 13 (10.5% Exon 21 c.2590C>T* (c.2589C>T) p.Arg864X* Mixed European 12, 28, 49 alleles12) Exon 22 c.2728C>T p.Arg910X 1 Italian 44
Exon 23 c.2929C>T p.Arg977X 1 Italian 44
Exon 26 c.3297G>A* p.Trp1099X* 2 Asian Indian Novel
Exon 27 c.3444C>Ga p.Tyr1148Xa 1 Japanese 24
Exon 28 c.3613C>T p.Gln1205X 2 Turkish 48
Exon 28 c.3652C>Ta p.Arg1218Xa 3 Mediterranean 20 3 (5.2% of Exon 28 c.3682C>T p.Arg1228X Caucasian 12 alleles12) 19 (26% of Turkish, Mixed Exon 31 c.3980G>A*a p.Trp1327X*a Turkish European, North 21, 27-29, 48 alleles48) African Exon31 c.4126C>T p.Gln1376X 6 Turkish 48
Exon 32 c.4193G>Aa p.Trp1398Xa 2 Mediterranean 20
MISSENSE MUTATIONS
Exon 5 c.413G>A* p.Gly138Glu* 1 Mixed European Novel
Exon 7 c.700T>Ca p.Cys234Arg 2 Italian 21
Exon 7 c.757G>C* p.Ala253Pro* 1 Mixed European Novel
Exon 11 c.1199T>C* p.Leu400Pro* 2 Caucasian Novel
Exon 11 c.1283G>A (c.1282G>A) p.Arg428Lys 1 Italian 49
Exon 13 c.1481G>A*b p.Arg494His*b 1 Caucasian Novel
Exon 13 c.1571G>A p.Arg524His 1 Italian 44
Exon 15 c.1859T>C p.Leu620Pro 2 Egyptian 32, 50 Exon 16 Not reported p.Gly655Arg 1 Caucasian 39
Exon 17 c.2023C>Ta p.Arg675Trpa 1 Tunisian 21
Exon 25 c.3259G>C (c.3258G>C) p.Gly1087Arg 2 Tunisian 49
Exon 26 c.3358G>C p.Ala1120Pro 1 Italian 44
Exon 27 c.3439A>G p.Arg1147Gly 2 Turkish 32
Exon 31 c.4090G>C p.Asp1364His 1 Korean 47
Exon 33 c.4342G>Ca p.Gly1448Arga 2 Japanese 51
Exon 35 c.4543T>C* p.Cys1515Arg* 1 Mixed European Novel
INTRONIC SPLICING MUTATIONS Disrupts donor splice Intron 4 c.293+1delG* 2 Mixed European Novel site* Skipping of exon 4, out Intron 4 c.293+2T>A (IVS4+2T>A) 4 Italian, Turkish 30, 48 of frame Disrupts donor splice Intron 4 c.293+2T>G Turkish 48 site Skipping of exon 6, in Intron 5 c.294-2A>C (IVS5-2A>C) 1 Japanese 52 frame Skipping of exon 6, in Intron 6 c.664+3A>G (IVS6+3A>G) 7 Italian 21, 44 frame 4 (2 Disrupts donor splice Intron 7 c.846+5G>A (IVS7+5G>A) homozygous Turkish 28 site sibs) Disrupts donor splice Intron 8 c.958+1G>A 2 Turkish 48 site c.1735+1G>T* Skipping exon 14, out Japanese, Intron 14 7 17, 35, 36, 43 (IVS14+1G>T) of frame* Chinese, Korean Disrupts acceptor Intron 18 c.2309-1G>A* 4 Mixed European Novel splice site* Japanese, c.2681+1G>A* Skipping exon 21, in 16 (23% of German, 20-22, 28, 30, Intron 21 (IVS21+1G>A) frame* alleles22) Mediterranean, 31 Asian Indian Disrupts donor splice Intron 21 c.2681+1G>T* 4 Mexican Novel site* c.2681+5insA Disrupts donor splice Intron 21 2 Turkish 28 (IVS21+5insA) site Disrupts donor splice Southeast Intron 25 c.3259+3A>T* 2 Novel site* Asian c.3362+1G>C Skipping exon 26, out Intron 26 2 Italian 30 (IVS26+1G>C) of frame c.3837-1G>A Activation of cryptic Intron 29 2 (sibs) Japanese 45 (IVS29-1G>A) splice site c.3837-17T>A Possible activation of Intron 29 1 Caucasian 39 (IVS29-17T>A) cryptic splice site c.4161+5G>A Intron 31 Unknown 1 Caucasian 39 (IVS31+5G>A) Japanese, c.4260-12A>G* Insertion of 11bp, 15 (5.5% of Intron 32 Chinese, 13, 24, 25 (IVS32-12A>G) p.1436X* alleles13) Caucasian Skipping exon 33> out c.4347+5G>A of frame> 1439X; Intron 33 1 Chinese 53 (IVS 33+5G>A) activation of cryptic site> p.Asp1421_Arg1467del SMALL DUPLICATION, INSERTION, AND DELETION FRAMESHIFT MUTATIONS c.18_19delGA 74.3% of GSD Exon 3 p.Gln6HisfsX20* Caucasian 12 (c.17_18delAG)* IIIb alleles4 p.Arg63ValfsX45 Exon 4 c.187delC 1 Japanese 45 (p.107X)
Exon 4 c.276delGa p.Gln92HisfsX16 1 Mediterranean 20
Exon 5 c.410_413delTGGG* p.Leu137TyrfsX20* 1 Mixed European Novel
Exon 5 c.442delA p.Arg148GlyfsX10 2 Italian 44
Exon 6 c.555_556delTT* p.Ser186LysfsX3* 1 Caucasian Novel
Exon 7 c.750-753delAGAC p.Asp251GlufsX23 2 Egyptian 27
Exon 7 c.753-756delGACA p.Asp251GlufsX23 1 Italian 44
Exon 7 c.672dupT (c.672insT)a p.Ser225Xa 2 (sibs) Mediterranean 20 Caucasian, Exon 9 c.1019delA p.Glu340AspfsX9 6 28, 48 Turkish
Exon 10 c.1117-1120delTCAA Exon 11 skipping 2 Unknown 54 Northern Exon 11 c.1276delG* p.Val426LeufsX12* 1 European/ Novel Mexican-Spanish Exon 12 c.1384delG* p.Val462X* 4 Hispanic Novel
c.1391dupG (c.1389insG; Egyptian, Exon 12 p.Asp465Arg 1 50, 55 c.1391-1392insG) Korean p.Thr558AsnfsX4 Exon 14 c.1672insAa 1 Japanese 45 (p.561X) a
Exon 16 c.1859-1869del 11bp p.Leu620HisfsX4 2 Iran 28
p.Gln667ArgfsX63 Exon 16 c.1999delCa 1 Japanese 53 (p.729X)a
Exon 17 c.2147delGa p.Gly716ValfsX14a 1 Italian 21
Exon 18 c.2223-2224delGT p.Gln741HisfsX7 1 Egyptian 50
Exon 19 c.2368dupA (c.2368insA) p.Arg790LysfsX4 2 Egyptian 50
Exon 21 c.2673delT p.Phe892LeufsX8 2 Egyptian 27
p.Gln906ProfsX5 Exon 22 c.2715-2719delTCAGA 2 Chinese 26 (p.910X)
Exon 23 c.2894_2896delGGAinsTG p.Trp965LeufsX2 1 Korean 47
p.Cys1005PhefsX7 Exon 24 c.3014delG* 1 Caucasian 13 (p.1011X)*
Exon 24 c.3049dupA* p.Thr1017AsnfsX53* 1 Caucasian Novel
Exon 25 c.3110dupT* p.Lys1038GlufsX32* 2 Asian Indian Novel
Exon 25 c.3144_3145delTG* p.Cys1048TrpfsX21* 2 Asian Indian Novel
Exon 25 c.3202_3203delTA p.Tyr1068X 2 Afghan 28
Exon 25 c.3216_3217delGA* p.Glu1072AspfsX36* 1 Unknown Novel African- Exon 27 c.3554delC* p.Thr1185LeufsX42* 1 Novel American p.Ala1222ValfsX5 Exon 28 c.3665delC 1? Unknown 13 (p.1226X)
Exon 28 c.3678_3681delAGAT* p.Asp1227GlufsX3* 2 Asian Indian Novel
p.Gly1273AsnfsX18 Exon 29 c.3816_3817delAGa 3 Japanese 45, 46 (p.1290X) p.Arg1279GlufsX11 Exon 29 c.3834delAa 2 Japanese 52 (p.1289X) a c.3911dupA (c.3911insA; p.Asn1304LysfsX7 Ashkenazi Exon 30 2 44, 49, 56 c.3904insA)c (p.1310X)c Jewish, Italian
Exon 31 c.3963delG p.Val1322SerfsX27 1 Italian 44 African p.Val1322AlafsX27 8 (6.7% of Exon 31 c.3965delT (c.3964delT) American, 13 (p.1348X) alleles13) Caucasian Exon 31 c.4043delT (c.4041delT) p.Leu1348X 2 Egyptian 50
p.Leu1408IlefsX14 Exon 32 c.4221dupA (c.4214insA) 1 Ashkenazi Jew 56 (p.1421X)
Exon 33 c.4324insAAa p.Gly1442LysfsX28a 3 Mediterranean 20 Mixed European/America Exon 33 c.4422delT* p.Ala1475GlnfsX4* 1 n Indian Novel
p.Ser1486ProfsX18 North African Exon 34 c.4455delT 100% 10 (p.1503X) Jews p.Tyr1510X Exon 35 c.4529dupA (c.4529insA)* 4 Caucasian 12 (p.Tyr1510X)* SMALL INFRAME DELETIONS AND INSERTIONS
Exon 12 Not reported p.delVal491_Arg494 1 Caucasian 29
Exon 16 c.1939_1941delTTG p.Val647del 1 Unknown 13
Exon 33 c.4335insTATa p.Tyr1445insa 1 Japanese 45
LARGE DELETIONS, DUPLICATIONS AND COMPLEX REARRANGEMENTS African- Exon 3 g.Exon 3 deletion* Exon 3 deletion* 2 Novel American Exon 9 c.966dup68*b Duplication/Frameshift* 2 Caucasian Novel c.3481- Exon 27 No mRNA detected 2 Egyptian 50 3588+1417del1525bp c.3512_3549dup + Amino acid Exon 27 1 Italian 49 3512_3519del rearrangement * - Mutations found in our study population. a – The nucleotide position for these mutations was originally reported to include 400bp 5’UTR, upstream from the ATG start codon. b - Variant found in study subjects with two known pathogenic mutations. c- Same mutation but with different nomenclature used for nucleotide position in different publications. ( )- If new nomenclature has been used to describe a specific mutation, parentheses indicate original nomenclature used when the mutation was initially reported.
REFERENCES 1. Gordon RB, Brown DH, Brown BI. Preparation and properties of the glycogendebranching enzyme from rabbit liver. Biochim Biophys Acta. Nov 10 1972;289(1):97-107. 2. Bates EJ, Heaton GM, Taylor C, Kernohan JC, Cohen P. Debranching enzyme from rabbit skeletal muscle; evidence for the location of two active centres on a single polypeptide chain. FEBS letters. Oct 15 1975;58(1):181-185. 3. Liu W, Madsen NB, Braun C, Withers SG. Reassessment of the catalytic mechanism of glycogen debranching enzyme. Biochemistry. Feb 5 1991;30(5):1419-1424. 4. Shen JJ, Chen YT. Molecular characterization of glycogen storage disease type III. Curr Mol Med. Mar 2002;2(2):167-175. 5. Ding JH, de Barsy T, Brown BI, Coleman RA, Chen YT. Immunoblot analyses of glycogen debranching enzyme in different subtypes of glycogen storage disease type III. J Pediatr. Jan 1990;116(1):95-100. 6. Van Hoof FaH, H.G. The subgroups of type III glycogenosis. European Journal of Biochemistry. 1967;2. 15 7. Yang BZ, Ding JH, Enghild JJ, Bao Y, Chen YT. Molecular cloning and nucleotide sequence of cDNA encoding human muscle glycogen debranching enzyme. J Biol Chem. May 5 1992;267(13):9294-9299. 8. Bao Y, Yang BZ, Dawson TL, Jr., Chen YT. Isolation and nucleotide sequence of human liver glycogen debranching enzyme mRNA: identification of multiple tissue-specific isoforms. Gene. Sep 15 1997;197(1-2):389-398. 9. Bao Y, Dawson TL, Jr., Chen YT. Human glycogen debranching enzyme gene (AGL): complete structural organization and characterization of the 5' flanking region. Genomics. Dec 1 1996;38(2):155-165. 10. Parvari R, Moses S, Shen J, Hershkovitz E, Lerner A, Chen YT. A single-base deletion in the 3'-coding region of glycogen-debranching enzyme is prevalent in glycogen storage disease type IIIA in a population of North African Jewish patients. Eur J Hum Genet. Sep-Oct 1997;5(5):266-270. 11. Santer R, Kinner M, Steuerwald U, et al. Molecular genetic basis and prevalence of glycogen storage disease type IIIA in the Faroe Islands. Eur J Hum Genet. May 2001;9(5):388-391. 12. Shen J, Bao Y, Liu HM, Lee P, Leonard JV, Chen YT. Mutations in exon 3 of the glycogen debranching enzyme gene are associated with glycogen storage disease type III that is differentially expressed in liver and muscle. J Clin Invest. Jul 15 1996;98(2):352-357. 13. Shaiu WL, Kishnani PS, Shen J, Liu HM, Chen YT. Genotype-phenotype correlation in two frequent mutations and mutation update in type III glycogen storage disease. Mol Genet Metab. Jan 2000;69(1):16-23. 14. Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. Sep 1 2002;30(17):3894-3900. 15. Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073-1081. 16. Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. Jul 1 2003;31(13):3812-3814. 17. Antonarakis SE. Recommendations for a nomenclature system for human gene mutations. Nomenclature Working Group. Hum Mutat. 1998;11(1):1-3. 18. den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat. 2000;15(1):7-12. 19. Shapiro MB, Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. Sep 11 1987;15(17):7155-7174. 20. Lucchiari S, Fogh I, Prelle A, et al. Clinical and genetic variability of glycogen storage disease type IIIa: seven novel AGL gene mutations in the Mediterranean area. Am J Med Genet. May 1 2002;109(3):183-190. 21. Lucchiari S, Donati MA, Parini R, et al. Molecular characterisation of GSD III subjects and identification of six novel mutations in AGL. Hum Mutat. Dec 2002;20(6):480. 22. Lucchiari S, Santoro D, Pagliarani S, Comi GP. Clinical, biochemical and genetic features of glycogen debranching enzyme deficiency. Acta Myol. Jul 2007;26(1):72-74. 23. Aoyama Y, Ozer I, Demirkol M, et al. Molecular features of 23 patients with glycogen storage disease type III in Turkey: a novel mutation p.R1147G associated with isolated glucosidase deficiency, along with 9 AGL mutations. J Hum Genet. Oct 16 2009. 16 24. Horinishi A, Okubo M, Tang NL, et al. Mutational and haplotype analysis of AGL in patients with glycogen storage disease type III. J Hum Genet. 2002;47(2):55- 59. 25. Okubo M, Horinishi A, Nakamura N, et al. A novel point mutation in an acceptor splice site of intron 32 (IVS32 A-12-->G) but no exon 3 mutations in the glycogen debranching enzyme gene in a homozygous patient with glycogen storage disease type IIIb. Hum Genet. Jan 1998;102(1):1-5. 26. Lam CW, Lee AT, Lam YY, et al. DNA-based subtyping of glycogen storage disease type III: mutation and haplotype analysis of the AGL gene in Chinese. Mol Genet Metab. Nov 2004;83(3):271-275. 27. Endo Y, Fateen E, Aoyama Y, et al. Molecular characterization of Egyptian patients with glycogen storage disease type IIIa. J Hum Genet. 2005;50(10):538- 542. 28. Endo Y, Horinishi A, Vorgerd M, et al. Molecular analysis of the AGL gene: heterogeneity of mutations in patients with glycogen storage disease type III from Germany, Canada, Afghanistan, Iran, and Turkey. J Hum Genet. 2006;51(11):958-963. 29. Schoser B, Glaser D, Muller-Hocker J. Clinicopathological analysis of the homozygous p.W1327X AGL mutation in glycogen storage disease type 3. Am J Med Genet A. Nov 15 2008;146A(22):2911-2915. 30. Hadjigeorgiou GM, Comi GP, Bordoni A, et al. Novel donor splice site mutations of AGL gene in glycogen storage disease type IIIa. J Inherit Metab Dis. Aug 1999;22(6):762-763. 31. Uotani S, Yamasaki H, Takino H, et al. Identification of a 5' splice junction mutation in the debranching enzyme gene in a Japanese patient with glycogen storage disease type IIIa. J Inherit Metab Dis. Jul 2000;23(5):527-528. 32. Cheng A, Zhang M, Okubo M, Omichi K, Saltiel AR. Distinct mutations in the glycogen debranching enzyme found in glycogen storage disease type III lead to impairment in diverse cellular functions. Hum Mol Genet. Jun 1 2009;18(11):2045-2052. 33. Shen J, Bao Y, Chen YT. A nonsense mutation due to a single base insertion in the 3'-coding region of glycogen debranching enzyme gene associated with a severe phenotype in a patient with glycogen storage disease type IIIa. Hum Mutat. 1997;9(1):37-40. 34. Labrune P, Trioche P, Duvaltier I, Chevalier P, Odievre M. Hepatocellular adenomas in glycogen storage disease type I and III: a series of 43 patients and review of the literature. J Pediatr Gastroenterol Nutr. Mar 1997;24(3):276-279. 35. Matern D, Starzl TE, Arnaout W, et al. Liver transplantation for glycogen storage disease types I, III, and IV. Eur J Pediatr. Dec 1999;158 Suppl 2:S43-48. 36. Shimizu J, Shiraishi H, Sakurabayashi S, et al. [A report on an adult case of type III glycogenosis with primary liver cancer and liver cirrhosis]. Nippon Shokakibyo Gakkai Zasshi. Dec 1982;79(12):2328-2332. 37. Siciliano M, De Candia E, Ballarin S, et al. Hepatocellular carcinoma complicating liver cirrhosis in type IIIa glycogen storage disease. J Clin Gastroenterol. Jul 2000;31(1):80-82. 38. Haagsma EB, Smit GP, Niezen-Koning KE, Gouw AS, Meerman L, Slooff MJ. Type IIIb glycogen storage disease associated with end-stage cirrhosis and hepatocellular carcinoma. The Liver Transplant Group. Hepatology. Mar 1997;25(3):537-540. 17 39. Demo E, Frush D, Gottfried M, et al. Glycogen storage disease type IIIhepatocellular carcinoma a long-term complication? J Hepatol. Mar 2007;46(3):492-498. 40. Cosme A, Montalvo I, Sanchez J, et al. [Type III glycogen storage disease associated with hepatocellular carcinoma]. Gastroenterol Hepatol. Dec 2005;28(10):622-625. 41. Silva AL, Romao L. The mammalian nonsense-mediated mRNA decay pathway: to decay or not to decay! Which players make the decision? FEBS letters. Feb 4 2009;583(3):499-505. 42. Liu HX, Cartegni L, Zhang MQ, Krainer AR. A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nat Genet. Jan 2001;27(1):55-58. 43. Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. Apr 2002;3(4):285-298. 44. Lucchiari S, Pagliarani S, Salani S, et al. Hepatic and neuromuscular forms of glycogenosis type III: nine mutations in AGL. Hum Mutat. Jun 2006;27(6):600- 601. 45. Okubo M, Horinishi A, Takeuchi M, et al. Heterogeneous mutations in the glycogen-debranching enzyme gene are responsible for glycogen storage disease type IIIa in Japan. Hum Genet. Jan 2000;106(1):108-115. 46. Ogimoto A, Okubo M, Okayama H, et al. A Japanese patient with cardiomyopathy caused by a novel mutation R285X in the AGL gene. Circ J. Oct 2007;71(10):1653-1656. 47. Oh SH, Park HD, Ki CS, Choe YH, Lee SY. Biochemical and molecular investigation of two Korean patients with glycogen storage disease type III. Clin Chem Lab Med. 2008;46(9):1245-1249. 48. Aoyama Y, Ozer I, Demirkol M, et al. Molecular features of 23 patients with glycogen storage disease type III in Turkey: a novel mutation p.R1147G associated with isolated glucosidase deficiency, along with 9 AGL mutations. J Hum Genet. Nov 2009;54(11):681-686. 49. Lucchiari S, Donati MA, Melis D, et al. Mutational analysis of the AGL gene: five novel mutations in GSD III patients. Hum Mutat. Oct 2003;22(4):337. 50. Endo Y, Fateen E, El Shabrawy M, et al. Egyptian glycogen storage disease type III - identification of six novel AGL mutations, including a large 1.5 kb deletion and a missense mutation p.L620P with subtype IIId. Clin Chem Lab Med. 2009;47(10):1233-1238. 51. Okubo M, Kanda F, Horinishi A, et al. Glycogen storage disease type IIIa: first report of a causative missense mutation (G1448R) of the glycogen debranching enzyme gene found in a homozygous patient. Hum Mutat. Dec 1999;14(6):542- 543. 52. Fukuda T, Sugie H, Ito M. Novel mutations in two Japanese cases of glycogen storage disease type IIIa and a review of the literature of the molecular basis of glycogen storage disease type III. J Inherit Metab Dis. Mar 2000;23(2):95-106. 53. Okubo M, Horinishi A, Suzuki Y, Murase T, Hayasaka K. Compound heterozygous patient with glycogen storage disease type III: identification of two novel AGL mutations, a donor splice site mutation of Chinese origin and a 1-bp deletion of Japanese origin. Am J Med Genet. Jul 31 2000;93(3):211-214. 54. Sugie H, Fukuda T, Ito M, Sugie Y, Kojoh T, Nonaka I. Novel exon 11 skipping mutation in a patient with glycogen storage disease type IIId. J Inherit Metab Dis. Oct 2001;24(5):535-545. 18 55. Kim KO, Lee HJ, Choi JW, Eun JR, Choi JH. An adult case of glycogen storage disease type IIIa. Korean J Hepatol. Jun 2008;14(2):219-225. 56. Parvari R, Shen J, Hershkovitz E, Chen YT, Moses SW. Two new mutations in the 3' coding region of the glycogen debranching enzyme in a glycogen storage disease type IIIa Ashkenazi Jewish patient. J Inherit Metab Dis. Apr 1998;21(2):141-148.