Table S1.Summary of number of environments, genotypes and markers used in this study. Breeding population Year Environments Total genotypes No. of DArT markers polymorphic 1 2009 3 368 1,411 2 2011 4 155 1,159
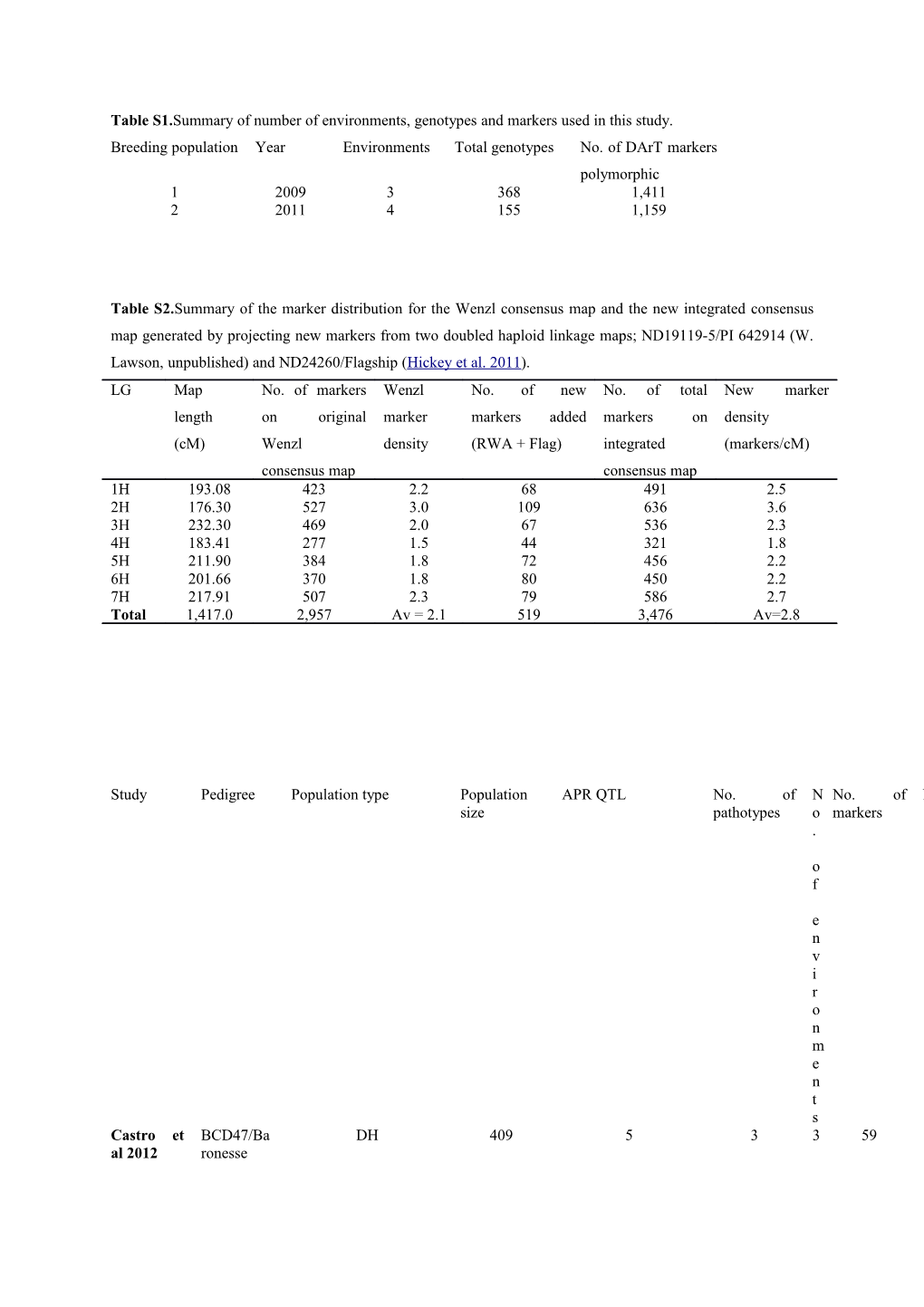
Table S2.Summary of the marker distribution for the Wenzl consensus map and the new integrated consensus map generated by projecting new markers from two doubled haploid linkage maps; ND19119-5/PI 642914 (W. Lawson, unpublished) and ND24260/Flagship (Hickey et al. 2011). LG Map No. of markers Wenzl No. of new No. of total New marker length on original marker markers added markers on density (cM) Wenzl density (RWA + Flag) integrated (markers/cM) consensus map consensus map 1H 193.08 423 2.2 68 491 2.5 2H 176.30 527 3.0 109 636 3.6 3H 232.30 469 2.0 67 536 2.3 4H 183.41 277 1.5 44 321 1.8 5H 211.90 384 1.8 72 456 2.2 6H 201.66 370 1.8 80 450 2.2 7H 217.91 507 2.3 79 586 2.7 Total 1,417.0 2,957 Av = 2.1 519 3,476 Av=2.8
Study Pedigree Population type Population APR QTL No. of N No. of No. of linkage groups size pathotypes o markers .
o f
e n v i r o n m e n t s Castro et BCD47/Ba DH 409 5 3 3 59 al 2012 ronesse Gonzalez L94/Vada RIL 103 - 3 1 957 et al 2012 L94/116-5 RIL 117 - 3 1 280 CebadaCap RIL 113 - 3 1 494 a/SusPtrit
Hickey et ND24260/ DH 334 8 2 5 1500 al 2011 Flagship
Kichereret Krona/HO DH 220 3 1 2 150 al 2000 R 1063
Liu et al Pompadour DH 292 1 Natural 5 514 2011 /Stirling infection
Marcel et Steptoe/Mo DH 150 - 1 1 635 al 2007 rex Oregon DH 94 - 1 1 797 Wolfe Barley Qi et al L94/Vada RIL 103 1* 3 1 2 1998
Von Korff Scarlett/IS BC2DH 301 6 1 4 98 et al 2005 R42-8
*Number of QTL detected in both plant stages Table S3.Details of the 8 QTL publications included in this study including pedigree, population type (DH: double haploid, RIL: recombinant inbred line, BC2DH: backcross two doubled haploid), population size (n), number of seedling QTL, number of APR QTL, total number of markers, total number of linkage groups detected, total map length (cM), marker density (average distance between markers), analysis method used (IM: interval mapping, CIM: composite interval mapping, MQM: multiple QTL method, MIM: multiple interval mapping).