TERMINAL FLOWER1 is a Mobile Signal Controlling Arabidopsis Architecture
Lucio Conti and Desmond Bradley
Supplemental Data
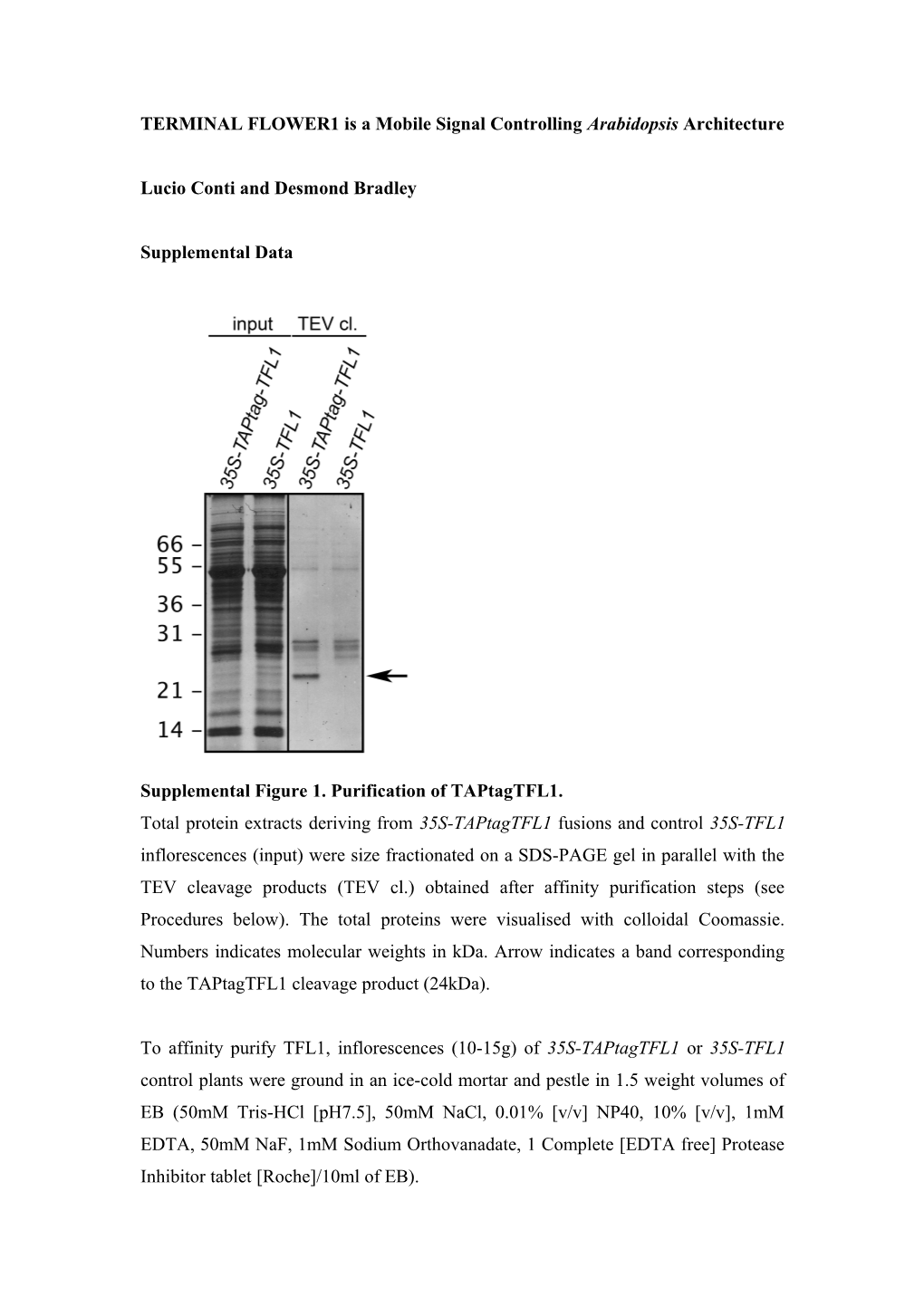
Supplemental Figure 1. Purification of TAPtagTFL1. Total protein extracts deriving from 35S-TAPtagTFL1 fusions and control 35S-TFL1 inflorescences (input) were size fractionated on a SDS-PAGE gel in parallel with the TEV cleavage products (TEV cl.) obtained after affinity purification steps (see Procedures below). The total proteins were visualised with colloidal Coomassie. Numbers indicates molecular weights in kDa. Arrow indicates a band corresponding to the TAPtagTFL1 cleavage product (24kDa).
To affinity purify TFL1, inflorescences (10-15g) of 35S-TAPtagTFL1 or 35S-TFL1 control plants were ground in an ice-cold mortar and pestle in 1.5 weight volumes of EB (50mM Tris-HCl [pH7.5], 50mM NaCl, 0.01% [v/v] NP40, 10% [v/v], 1mM EDTA, 50mM NaF, 1mM Sodium Orthovanadate, 1 Complete [EDTA free] Protease Inhibitor tablet [Roche]/10ml of EB). The crude homogenate was clarified by spinning at 2800g at 4°C for 10min. The supernatant was filtered through 2 layers of Miracloth and spun again. The supernatant was incubated at 4°C with rotation for 1-2h with IgG beads slurry (Amersham-Pharmacia) pre-equilibrated in Buffer 1 (10mM Tris-HCl [pH 7.5], 100mM NaCl, 0.1% [v/v] NP40, 10% [v/v] Glycerol, 0.5mM EDTA). The beads were recovered and washed 3 times with ice-cold Buffer 1. The beads were then equilibrated in TEV buffer (10mM Tris-HCl [pH 8], 100mM NaCl, 0.1% [v/v] NP40, 0.5mM EDTA, 1mM DTT) and transferred to an Eppendorf tube in a total volume of 500µl of fresh TEV buffer. The tubes were mixed with TEV protease (Invitrogen) (1U/ 5mg of total protein extract), and incubated at 17°C for 2-3h. The TEV cleavate was recovered and concentrated 10 fold by adding StrataClean resin (Stratagene), before running on a 12% SDS-PAGE gel and staining for total protein with Colloidal Coomassie G250. A band corresponding to the TEV cleavate product of TAPtagTFL1 (24kDa) was processed for MALDI-TOF at the JIC proteomic facility. The tryptic digest was analysed with Mascot (www.matrixscience.com) and FindMod (www.expasy.org/tools/findmod) set with a mass tolerance of 50 ppm (equivalent to 0.005% Da). Arabidopsis plants (tfl1-1 background) were transformed with pLC37 via Agrobacterium (Clough, S.J. and Bent, A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735- 743). Independent single-insertion homozygous plants having a functional 35S::TAPtag-TFL1 transgene were isolated through Basta selection.