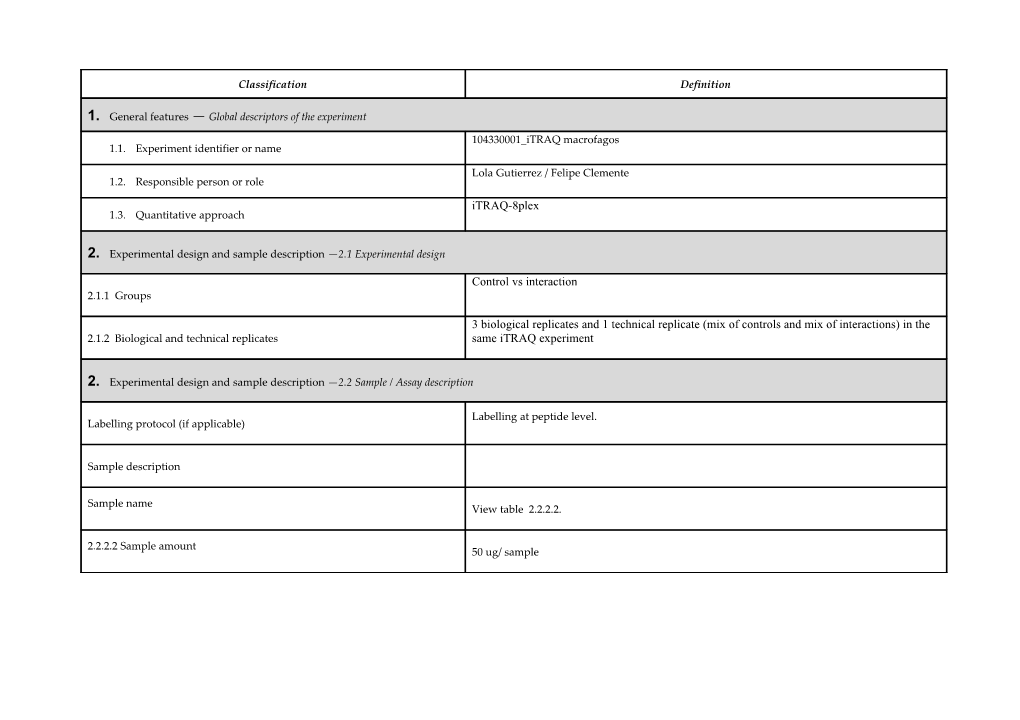
Classification Definition
1. General features — Global descriptors of the experiment
104330001_iTRAQ macrofagos 1.1. Experiment identifier or name
Lola Gutierrez / Felipe Clemente 1.2. Responsible person or role iTRAQ-8plex 1.3. Quantitative approach
2. Experimental design and sample description —2.1 Experimental design
Control vs interaction 2.1.1 Groups
3 biological replicates and 1 technical replicate (mix of controls and mix of interactions) in the 2.1.2 Biological and technical replicates same iTRAQ experiment
2. Experimental design and sample description —2.2 Sample / Assay description
Labelling at peptide level. Labelling protocol (if applicable)
Sample description
Sample name View table 2.2.2.2.
2.2.2.2 Sample amount 50 ug/ sample Sample name iTRAQ reagent
CONTROL 1 113
2.2.2.3 Sample labelling with assay definition, i.e. MS run / data set together CONTROL 3 114 with reporting ion mass, reagent or isotope labelled amino acid CONTROL 4 115 INTERACCION 1 116 INTERACCION 3 117 INTERACCION 4 118 CONTROL MIX 119 INTERACCION MIX 121
2.2.2.4 Replicates and/or groups View table 2.2.2.2.
Reagent % of -2 % of -1 % of 0 % of +1 % of +2 iTRAQ113 0.00 0.00 92.87 6.89 0.24 iTRAQ114 0.00 0.94 93.00 5.90 0.16 iTRAQ115 0.00 1.88 93.12 4.90 0.10 iTRAQ116 0.00 2.82 93.21 3.90 0.07 2.2.3 Isotopic correction coefficients iTRAQ117 0.06 3.77 93.29 2.88 0.00 iTRAQ118 0.09 4.71 93.29 1.91 0.00 iTRAQ119 0.14 5.66 93.33 0.87 0.00 iTRAQ121 0.27 7.44 92.11 0.18 0.00
List of internal references used and their amount. Also state their specific purpose such as 2.2.4 Internal references quantification, normalization or alignment.
3 Input data — Description and reference of the dataset used for quantitative analysis: type, format and availability of the data. No actual values are requested here.
3.1 Input data type tandem MS
Import from oracle DB 4000 Series Explorer v 3.6 3.2 Input data format 4800 MALDI TOF/TOF ABI Sciex ( .t2d). Sample was fractionated using a 3100 off-gel fractionator (Agilent Technologies), collecting 24 fractions on pH range of 3-10. Each fraction was individually fractionated in a nano-HPLC (LC Packings), coupled off line to a 4800 Plus MALDI-TOF/TOF (ABSciex), each off-gel 3.3 Input data merging fraction yields 416 MALDI spots. Datasets corresponding to each fraction were merged and combined by the Protein Pilot software v3.0 (ABSciex) for identification and quantitation purposes.
3.4 Availability of the input data http://proteo.cnb.csic.es/downloads/miape-quant/iTRAQ8plex_UCMPCM_peaklist.xml
4 Protocol —Description of the software and methods applied in the quantitative analysis (including transformation functions, aggregation functions and statistical calculations).
4.1 Quantification software name, version and manufacturer Protein Pilot v.3.0 from AbSciex
4.2 Description of the selection and/or matching method of features, Centroid, peak area and peak area error. together with the description of the method of the primary extracted Ratio of peak reporter area, the protein ratio is calculated as average ratio: quantification values determination for each feature and/or peptide 10exp(weighted average of log ratios)/bias Peptide confidence is based on the score, which is the number of matches between the data and the theoretical fragment ions. In general, a higher score leads to a higher confidence, but the factors listed below also influence the confidence. Hypothesized modifications – Rare, unexpected modifications tend to decrease confidence. Delta mass – A larger delta mass tends to decrease confidence. 4.3 Confidence filter of features or peptides prior to quantification Peptide cleavage – Cleavages inconsistent with the digest agent specified in the analysis method tend to decrease confidence. Alternative hypotheses for the same spectrum – A peptide with a high score could receive a low confidence because there is another peptide hypothesis for this spectrum with an even higher score.
4.4 Description of data calculation and transformation methods
Peptides with a combined feature probability < 30% are excluded . Features with this low probability include semi-tryptic peptides, peptides missing an iTRAQ reagent 4.4.1 Missing values imputation and outliers removal label, peptides with low probability modifications and peptides with large delta masses. Peptide ratios 0 and 9999.99 4.4.2 Quantification values calculation and / or ratio determination from Ratios of peak area iTRAQ reporters. the primary extracted quantification values Normalization: mean protein iTRAQ ratio in all replicates and all ratios in each replicate 4.4.3 Replicate aggregation Are divided by mean. Average ratio for quantified protein in all replicates and p-value (<0.05) and/or ST < 0.2
4.4.4 Normalization Auto bias correction: median average protein ratio..
As stated in point 4.4. For shared peptides: A shared peptide is when the same spectrum with the same peptide sequence is claimed by more than one protein. These peptides are excluded when there is another protein which claims this peptide but only if the Unused ProtScore for the other protein is 1.3 or higher. 4.4.5 Protein quantification values calculation and / or ratio Precursor overlap – The spectrum yielding the identified peptide is also claimed determination from the peptide quantification values by a different protein above the confidence cutoff, but with an unrelated peptide sequence. This can happen when more than one peptide is fragmented at the same time. These peptides are excluded. Only proteins with at least two peptides identified are included in the quantification.
4.4.6 Protocol specific corrections
4.5 Description of methods for (statistical) estimation of correctness p-value < 0.05; EF<2; FDR 1%
4.6 Calibration curves of standards
5 Resulting data —Provide the actual quantification values resulting from your quantification software together with their estimated confidence. Depending of the quantification technique or even of the quantification software, only some of the following items could be satisfied (e.g., for spectral counting, only quantification values at protein level can be provided) 5.1 Quantification values at peptide and/or feature level: Actual quantification values achieved for each peptide and/or, in case of feature-based quantification, for the corresponding features (mapped back from each peptide), together with their estimated confidence.
5.1.1 Primary extracted quantification values for each feature (e.g. area, http://proteo.cnb.csic.es/downloads/miape-quant/iTRAQ8plex_UCMPCM_peptide summary.xls height, etc.), with their statistical estimation of correctness 5.1.2 Quantification values for each peptide as a result of the aggregation of the values of the previous section (5.1.1), with their statistical estimation of correctness 5.2 Quantification values at protein level: Actual quantification values achieved for each protein and for each protein ambiguity group, together with the confidence in the quantification value.
http://proteo.cnb.csic.es/downloads/miape-quant/iTRAQ8plex_UCMPCM_protein Basic / raw quantification values with statistical estimation of correctness summary.xls
5.2.2 Transformed / aggregated / combined quantification values of the http://proteo.cnb.csic.es/downloads/miape-quant/iTRAQ8plex_UCMPCM_significant proteins at group level, with their statistical estimation of correctness changes_R1R3Rmix.xls