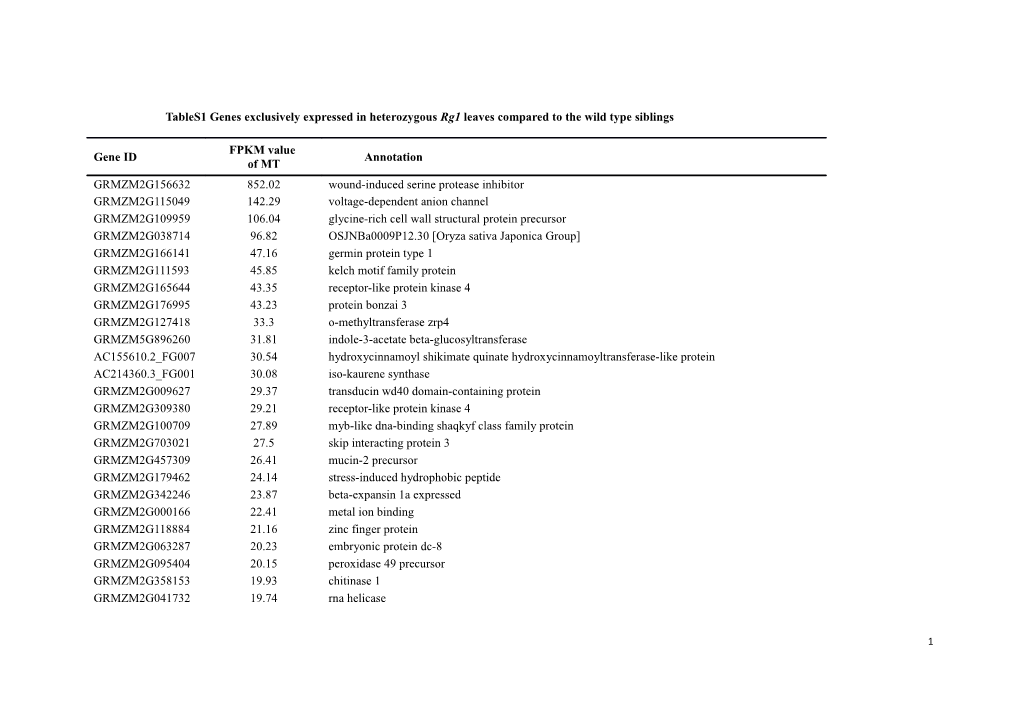
TableS1 Genes exclusively expressed in heterozygous Rg1 leaves compared to the wild type siblings
FPKM value Gene ID Annotation of MT GRMZM2G156632 852.02 wound-induced serine protease inhibitor GRMZM2G115049 142.29 voltage-dependent anion channel GRMZM2G109959 106.04 glycine-rich cell wall structural protein precursor GRMZM2G038714 96.82 OSJNBa0009P12.30 [Oryza sativa Japonica Group] GRMZM2G166141 47.16 germin protein type 1 GRMZM2G111593 45.85 kelch motif family protein GRMZM2G165644 43.35 receptor-like protein kinase 4 GRMZM2G176995 43.23 protein bonzai 3 GRMZM2G127418 33.3 o-methyltransferase zrp4 GRMZM5G896260 31.81 indole-3-acetate beta-glucosyltransferase AC155610.2_FG007 30.54 hydroxycinnamoyl shikimate quinate hydroxycinnamoyltransferase-like protein AC214360.3_FG001 30.08 iso-kaurene synthase GRMZM2G009627 29.37 transducin wd40 domain-containing protein GRMZM2G309380 29.21 receptor-like protein kinase 4 GRMZM2G100709 27.89 myb-like dna-binding shaqkyf class family protein GRMZM2G703021 27.5 skip interacting protein 3 GRMZM2G457309 26.41 mucin-2 precursor GRMZM2G179462 24.14 stress-induced hydrophobic peptide GRMZM2G342246 23.87 beta-expansin 1a expressed GRMZM2G000166 22.41 metal ion binding GRMZM2G118884 21.16 zinc finger protein GRMZM2G063287 20.23 embryonic protein dc-8 GRMZM2G095404 20.15 peroxidase 49 precursor GRMZM2G358153 19.93 chitinase 1 GRMZM2G041732 19.74 rna helicase
1 GRMZM2G021786 19.48 udp-glycosyltransferase-like protein GRMZM2G138468 19.35 alpha-amylase GRMZM2G061230 18.26 peroxidase GRMZM2G014508 17.97 geranyl diphosphate synthase GRMZM2G046750 17.55 lipid binding protein GRMZM2G416836 17.31 trehalose-6-phosphate synthase GRMZM2G073764 17.11 hypothetical protein LOC100303899 [Zea mays] GRMZM2G453571 17.09 wrky transcription GRMZM5G812228 16.73 acyl- synthetase GRMZM2G428040 16.39 transcription activator-related protein GRMZM2G130454 16.33 lipid transfer protein GRMZM2G138355 15.46 nudix hydrolase 13 GRMZM2G146380 15.22 nac domain protein nac5 GRMZM2G036290 15.01 1-deoxy-d-xylulose 5-phosphate reductoisomerase GRMZM2G005633 14.7 chitinase GRMZM2G147221 14.69 cellulase containing protein GRMZM2G363893 14.56 2og-fe oxygenase family expressed GRMZM2G326116 14.47 isoflavone expressed GRMZM2G155375 14.01 acyl- binding family expressed AC194970.5_FG002 13.33 protein brassinazole-resistant 2 GRMZM2G103137 13.03 hydrolase-like protein GRMZM2G152451 12.19 hypothetical protein LOC100275330 [Zea mays] GRMZM2G075255 12.03 cer1 protein GRMZM2G022318 11.93 heavy metal-associated domain containing expressed GRMZM2G141584 11.83 tartrate-resistant acid phosphatase type 5 precursor GRMZM2G094523 11.48 expansin os-expa3 GRMZM2G042933 11.23 amino acid permease GRMZM2G412436 10.78 late embryogenesis abundant domain-containing protein GRMZM2G012262 10.62 pbf-2 like transcription factor
2 GRMZM2G160473 10.55 dicer-like protein 4 GRMZM5G856011 10.46 serine threonine kinase protein GRMZM2G093146 10.14 dehydrodolichyl diphosphate synthase 6 GRMZM2G025190 10.11 glutathione s-transferase GRMZM2G465046 10.01 zinc finger GRMZM2G471065 9.82 mannan endo- -beta-mannosidase 6 GRMZM2G021427 9.8 beta-expansin 1a expressed GRMZM2G170420 9.48 40s ribosomal protein s4 GRMZM2G327266 9.45 beta-expansin 4 precursor GRMZM2G167673 9.39 cytochrome p450 GRMZM2G070588 9.16 nucleic acid binding GRMZM2G082249 8.98 cytokinin-o-glucosyltransferase 3 GRMZM2G070034 8.81 transcription factor mads56 GRMZM2G013398 8.8 cct motif family protein GRMZM2G434170 8.72 purple acid phosphatase GRMZM2G471670 8.69 heat shock -like GRMZM2G031790 8.36 fatty acid elongase GRMZM2G093125 8.35 tyrosine dopa decarboxylase GRMZM2G454889 8.23 hypothetical protein SORBIDRAFT_01g006230 [Sorghum bicolor] GRMZM2G126541 8.01 serine carboxypeptidase AC217910.3_FG001 7.91 metal ion binding protein GRMZM2G097851 7.88 blue copper protein GRMZM2G139617 7.78 dek domain-containing chromatin associated protein GRMZM2G031323 7.73 myb-related protein m4 GRMZM2G431018 7.7 serine threonine-specific protein kinase GRMZM2G162158 7.25 iron ascorbate-dependent oxidoreductase GRMZM5G812326 7.23 Os01g0389700 [Oryza sativa Japonica Group] GRMZM2G028134 7.02 metal ion binding protein GRMZM2G017629 6.97 jasmonate-induced protein
3 GRMZM2G142390 6.85 kinase-like protein GRMZM2G136146 6.85 hypothetical protein SORBIDRAFT_03g041085 [Sorghum bicolor] AC212835.3_FG008 6.83 protein kinase GRMZM2G376085 6.57 ring u-box domain and arm repeat-containing protein GRMZM2G147420 6.53 ulp1 protease c-terminal catalytic domain containing expressed GRMZM2G110324 6.52 lysine-ketoglutarate reductase saccharopine dehydrogenase bifunctional enzyme GRMZM2G404316 6.47 hypothetical protein LOC100304202 [Zea mays] GRMZM2G022793 6.22 phosphate phosphoenolpyruvate translocator GRMZM2G092925 6.05 hva22e GRMZM2G312910 5.96 shoot1 protein GRMZM2G043300 5.88 chemocyanin precursor GRMZM2G401521 5.87 wrky23 - superfamily of tfs having wrky and zinc finger domains GRMZM2G000601 5.74 ubiquitin-conjugating enzyme e2-17 kda GRMZM2G019974 5.59 organic cation transporter GRMZM2G145221 5.57 bzip protein GRMZM2G146518 5.45 pentatricopeptide repeat-containing GRMZM2G309731 5.42 ap2 erf domain-containing transcription factor GRMZM2G181030 5.39 at5g37260-like protein GRMZM2G099297 5.28 o-methyltransferase zrp4 GRMZM5G822426 5.11 tetratricopeptide repeat GRMZM2G174537 4.8 vesicle-associated membrane protein-associated protein a GRMZM2G005624 4.67 grassy tillers 1 GRMZM5G849099 4.61 hypothetical protein SORBIDRAFT_03g031430 [Sorghum bicolor] GRMZM2G082118 4.46 vq motif family protein GRMZM2G336337 4.45 laccase 90c GRMZM2G441031 4.36 wrky dna binding domain containing expressed GRMZM2G067919 4.36 globulin 1 GRMZM2G134687 4.19 nac transcription factor GRMZM2G171279 3.77 potassium channel
4 GRMZM2G147399 3.55 early nodulin 93 GRMZM2G056988 3.54 hypothetical protein LOC100277951 [Zea mays] GRMZM2G304049 3.51 lz-nbs-lrr class rga GRMZM2G062910 3.34 mterf family protein GRMZM2G156490 3.32 hypothetical protein LOC100382083 [Zea mays] GRMZM2G051921 3.3 chitinase GRMZM2G009465 3.27 proline-rich protein GRMZM2G156977 2.94 nac domain-containing protein 21 22 MT, mutant. FPKM: fragment per kilobase of exon per million fragments mapped.
TableS2 Significantly (P<0.05) differentially expressed genes with more than threefold expression in Rg1 leaves compared to the wild type siblings
FPKM FPKM value Fold change Gene value Annotation of MT MT/WT of WT GRMZM2G177726 2327.66 7.49 310.82 protein kinase GRMZM2G000221 259.38 2.17 119.47 pvr3-like protein GRMZM2G010048 80.2 0.68 117.91 p21 expressed GRMZM5G878346 92.67 1.11 83.81 hypothetical protein LOC100381757 [Zea mays] GRMZM2G053669 214.48 2.81 76.44 asparagine synthetase GRMZM2G102183 24.04 0.39 62.18 malate synthase AC148152.3_FG005 35.38 0.64 55.39 1-aminocyclopropane-1-carboxylate oxidase GRMZM2G424205 104.46 2.12 49.38 hypothetical protein [Zea mays] xyloglucan endotransglucosylase hydrolase protein GRMZM2G413044 42.12 0.98 43.2 32 precursor GRMZM2G056236 26.59 0.64 41.56 beta-expansin 4 precursor
5 GRMZM2G079583 28.29 0.68 41.55 serine threonine-specific protein kinase npk15-like GRMZM2G175406 83.25 2.06 40.51 pro-resilin precursor GRMZM2G126772 84.24 2.13 39.59 benzoate carboxyl methyltransferase GRMZM2G054332 13.01 0.33 39.23 white-brown-complex abc transporter family GRMZM2G119305 39.56 1.04 38.1 stearoyl-acyl carrier protein desaturase GRMZM2G007477 26.83 0.73 36.62 serine-threonine protein plant- GRMZM2G444560 21.9 0.64 34.15 aaa-type atpase-like GRMZM5G812272 22.07 0.69 31.8 wrky transcription factor 71 GRMZM2G113421 23.38 0.74 31.79 rust resistance kinase Lr10 GRMZM2G079440 166.8 5.34 31.26 dehydrin7 GRMZM2G028393 2815.27 92.04 30.59 maize proteinase inhibitor serine threonine protein phosphatase 2a regulatory GRMZM2G102858 51.12 1.68 30.44 subunit GRMZM2G059939 27.39 0.92 29.89 ap2 domain-containing transcription factor hydroxycinnamoyl shikimate quinate GRMZM2G034360 37.96 1.3 29.3 hydroxycinnamoyltransferase-like protein GRMZM2G125032 277.37 9.54 29.08 beta- -glucanase GRMZM2G078472 15.74 0.54 28.91 asparagine synthetase GRMZM2G013002 165.63 5.74 28.85 beta-expansin 1a expressed GRMZM2G099619 25.59 0.9 28.46 abc transporter family protein GRMZM2G082520 259.15 9.47 27.36 beta-expansin 1a expressed GRMZM2G122302 28.69 1.05 27.25 blue copper protein GRMZM2G002260 8.3 0.31 27.17 glycosyl hydrolase family 10 protein GRMZM2G004160 173.88 6.64 26.19 chemocyanin precursor GRMZM2G339122 346.26 13.25 26.13 expansin os-expa2 GRMZM2G004694 62.27 2.54 24.52 sorbitol transporter branched-chain-amino-acid aminotransferase GRMZM2G153536 23.32 0.97 23.98 chloroplast expressed GRMZM2G175761 21.21 0.91 23.42 pollen-specific protein sf3 GRMZM2G143204 23.41 1.04 22.49 wrky transcription factor 57
6 GRMZM2G145518 30.89 1.42 21.8 chitinase GRMZM2G041235 9.73 0.45 21.67 hypothetical protein LOC100216925 [Zea mays] GRMZM5G858417 10.05 0.47 21.5 permease 1 GRMZM2G075315 47.7 2.28 20.94 bowman-birk type trypsin inhibitor hypothetical protein SORBIDRAFT_01g030310 GRMZM2G327907 628.81 31.05 20.25 [Sorghum bicolor] GRMZM2G137535 620.52 31.42 19.75 lichenase-2 precursor wrky11 - superfamily of tfs having wrky and zinc AC198725.4_FG009 14.56 0.74 19.66 finger domains GRMZM2G024104 43.9 2.24 19.64 glutamine synthetase GRMZM2G105221 33.15 1.71 19.35 glycosyltransferase family 61 protein AC197758.3_FG004 49.91 2.63 18.99 peroxidase 52 GRMZM2G425798 46.12 2.47 18.66 hypothetical protein LOC100275603 [Zea mays] GRMZM5G898290 57.56 3.1 18.59 nac domain protein nac5 GRMZM2G106493 45.73 2.47 18.52 receptor-type protein kinase lrk1 GRMZM2G085967 78.62 4.28 18.35 peroxidase 3 GRMZM2G407044 27.61 1.54 17.96 acetolactate synthase amino acid binding protein GRMZM2G154007 40.18 2.24 17.92 alcohol dehydrogenase GRMZM2G117942 43.48 2.56 17.01 pathogenesis-related protein 4 GRMZM2G181605 20.14 1.2 16.81 nac domain ipr003441 GRMZM2G065585 14.97 0.9 16.57 beta glucanase GRMZM2G023081 20.91 1.27 16.52 pgps d12 GRMZM2G475380 78.28 4.93 15.87 2og-fe oxygenase family expressed GRMZM2G442387 15.61 1.01 15.4 binding protein GRMZM2G127379 20.66 1.35 15.28 nac domain transcription factor GRMZM2G052880 50.29 3.3 15.24 snare-interacting protein keule GRMZM2G051943 474.57 31.35 15.14 chitinase terpenoid cylase protein prenyltransferase alpha- GRMZM2G146981 20.57 1.36 15.09 alpha toroid GRMZM2G176595 224.2 15.03 14.92 beta-expansin 1a precursor
7 GRMZM2G042278 8.32 0.56 14.84 rwp-rk domain-containing protein GRMZM2G047139 54.85 3.71 14.78 esterase precursor GRMZM5G805609 50.43 3.43 14.72 glucan endo- -beta-glucosidase 7 precursor non-cell-autonomous protein pathway2 GRMZM2G041356 36.02 2.46 14.65 plasmodesmal receptor GRMZM2G178910 30.85 2.12 14.53 dna binding GRMZM2G059964 56.95 3.97 14.34 lipid transfer protein GRMZM2G466298 29.38 2.06 14.25 lectin-like protein kinase GRMZM2G025832 10.26 0.72 14.23 flavonoid 3 -hydroxylase GRMZM2G056989 144.98 10.41 13.93 retrotransposon expressed GRMZM2G052266 12.54 0.91 13.81 sarcosine oxidase GRMZM2G145045 10.33 0.75 13.79 wall-associated kinase 4 GRMZM2G355752 432.54 31.76 13.62 early light-induced protein GRMZM2G061806 10.14 0.75 13.5 transferase family expressed GRMZM2G107073 23.61 1.75 13.46 polygalacturonase-like protein GRMZM2G095126 185.65 13.89 13.36 alpha-galactosidase alpha-n- GRMZM2G109130 75.22 5.68 13.25 lipoxygenase 1 GRMZM2G152417 15.81 1.22 12.95 amp dependent GRMZM2G028821 45.9 3.61 12.73 glutathione-s-transferase GRMZM2G077914 13.38 1.06 12.58 rust resistance kinase Lr10 GRMZM2G122267 77.93 6.22 12.54 ph domain containing protein AC195347.4_FG004 12.84 1.03 12.47 f-box lrr-repeat GRMZM2G126900 275.47 22.09 12.47 myo-inositol oxygenase GRMZM2G177934 80.88 6.72 12.04 chemocyanin precursor GRMZM2G089102 11.49 0.96 11.94 act domain-containing protein 3 GRMZM2G075438 21.49 1.82 11.83 serine threonine kinase -related protein GRMZM2G034611 30.81 2.61 11.82 rust resistance kinase Lr10 GRMZM2G133802 93.33 7.92 11.79 beta chain GRMZM2G053779 81.72 7.01 11.65 blue copper protein
8 GRMZM2G428987 19.78 1.71 11.55 pir7b protein GRMZM2G039639 41.48 3.62 11.47 zeamatin-like protein AC193632.2_FG002 9.8 0.86 11.43 receptor serine threonine kinase pr5k-like GRMZM2G122447 86.29 7.6 11.35 vq motif family protein wapl (wings apart-like protein regulation of AC208440.3_FG003 20.45 1.81 11.29 heterochromatin) protein GRMZM2G133937 45.08 4.02 11.2 sac domain-containing protein 9 GRMZM2G470882 56.57 5.07 11.16 unknown [Zea mays] GRMZM2G160710 39.21 3.55 11.04 nbs-lrr class disease resistance protein GRMZM2G340656 560.39 50.94 11 stachyose synthase precursor GRMZM2G328785 7.03 0.64 10.96 receptor-like protein kinase GRMZM2G035131 36.49 3.33 10.95 lysophospholipase homolog GRMZM2G128929 13.44 1.24 10.84 lactate dehydrogenase GRMZM2G025924 5.32 0.49 10.84 histone-lysine n- h3 lysine-9 specific suvh9 GRMZM2G368886 14.47 1.35 10.73 alpha-expansin 6 precursor GRMZM2G005771 72.18 6.74 10.7 regulator of ribonuclease activity a GRMZM2G094510 45.24 4.27 10.61 hypothetical protein LOC100278059 [Zea mays] GRMZM5G813363 15.28 1.45 10.55 pollen specific protein GRMZM2G055802 35.39 3.4 10.42 bowman-birk type trypsin inhibitor GRMZM2G085924 11.53 1.11 10.37 o-methyltransferase family expressed loxc2_orysj ame: full=probable lipoxygenase GRMZM5G822593 11.79 1.14 10.34 chloroplastic flags: precursor GRMZM2G431288 7.31 0.71 10.33 elicitor-inducible cytochrome p450 senescence-inducible chloroplast stay-green GRMZM2G091837 13.24 1.28 10.32 protein 1 GRMZM2G354909 117.29 11.43 10.26 carbonyl reductase 3 GRMZM5G825854 8.25 0.81 10.12 sec34-like protein GRMZM2G085176 43.66 4.33 10.09 esterase precursor GRMZM2G101116 27.73 2.76 10.06 pirin-like protein GRMZM2G427097 25.91 2.59 10 glutamate dehydrogenase 2
9 GRMZM2G125441 47.06 4.71 9.99 meiosis 5 GRMZM2G373522 238.98 23.99 9.96 dehydrin cor410 GRMZM2G427815 17.06 1.71 9.95 peroxidase GRMZM2G349187 14.96 1.51 9.88 cytochrome p450 GRMZM2G083195 67.22 6.82 9.86 glycerol-3-phosphate acyltransferase 6 GRMZM2G302604 76.03 7.77 9.78 sorbitol transporter GRMZM2G119755 26.9 2.77 9.73 pgps d12 GRMZM5G817964 23.54 2.42 9.72 white-brown-complex abc transporter family GRMZM2G004795 63.63 6.57 9.69 ccch transcription factor GRMZM2G075456 538.31 55.78 9.65 calcium binding ef-hand protein GRMZM2G135165 18.31 1.91 9.61 hypothetical protein LOC100384836 [Zea mays] GRMZM2G332562 6.53 0.68 9.59 amino acid transporter GRMZM2G080183 8.71 0.91 9.58 peroxidase 16 GRMZM2G162359 40.22 4.29 9.39 class iii chitinase homologue ( hib3h-a) hypothetical protein SORBIDRAFT_10g024300 GRMZM2G132331 25.66 2.74 9.36 [Sorghum bicolor] GRMZM2G079219 24.17 2.58 9.35 rust resistance kinase Lr10 GRMZM2G412885 12.17 1.31 9.28 atp binding GRMZM2G029912 168.36 18.21 9.24 gl1 protein GRMZM2G076943 9.43 1.02 9.21 serine threonine kinase protein GRMZM2G126975 11.98 1.3 9.18 hypothetical protein LOC100192940 [Zea mays] GRMZM5G855337 17.29 1.9 9.1 ctp synthase GRMZM2G056500 106.81 11.74 9.09 pore-forming toxin-like protein hfr-2 brassinosteroid insensitive 1-associated receptor GRMZM2G104760 12.01 1.32 9.08 kinase 1 precursor GRMZM2G352855 101.64 11.21 9.07 acyltransferase ( kd) GRMZM2G145651 10.53 1.16 9.07 hydrolyzing o-glycosyl GRMZM2G044132 1344.66 148.92 9.03 abscisic stress ripening GRMZM2G076307 14.23 1.59 8.97 glycosyl transferase
10 GRMZM2G124759 1263.16 141.56 8.92 triacylglycerol lipase GRMZM2G178072 32.19 3.62 8.9 phd-finger protein GRMZM2G132303 26.87 3.04 8.83 dna-3-methyladenine glycosylase i GRMZM2G153799 31.58 3.58 8.82 polyamine oxidase GRMZM2G154958 10.78 1.22 8.81 proline transporter GRMZM2G159587 7.49 0.85 8.77 d-isomer specific 2-hydroxyacid dehydrogenase GRMZM2G122431 7.93 0.91 8.75 cellulose synthase isolog GRMZM2G011742 30.7 3.53 8.69 hydroxyproline-rich glycoprotein GRMZM2G145109 6.66 0.77 8.65 wall-associated kinase 4 GRMZM2G151468 12.71 1.51 8.42 beta-amylase GRMZM2G153087 11.2 1.34 8.36 phd finger protein GRMZM2G168115 21.66 2.6 8.32 aspartyl protease family protein GRMZM2G431039 52.03 6.27 8.3 beta- -glucanase GRMZM2G117989 110.5 13.34 8.29 win1 precursor GRMZM2G089163 16.19 1.95 8.28 dsrna-binding protein 3 GRMZM2G000620 46.28 5.6 8.26 receptor-like kinase GRMZM2G006964 11.77 1.43 8.21 gibberellin 2-beta-dioxygenase 7 GRMZM2G138450 10.34 1.26 8.2 peroxidase 27 GRMZM2G043600 66.1 8.06 8.2 dna binding protein GRMZM2G087326 38.76 4.74 8.18 reversibly glycosylated polypeptide GRMZM2G098890 7.94 0.97 8.17 glucosyl transferase GRMZM2G093272 44.58 5.46 8.17 usp family protein GRMZM2G083783 9.54 1.17 8.13 rna recognition motif-containing protein y2982_orysj ame: full=b3 domain-containing GRMZM5G834874 13.72 1.69 8.12 protein os02g0598200 GRMZM2G330302 28.64 3.55 8.07 ctp synthase GRMZM2G113821 131.99 16.43 8.03 ubiquitin carboxyl-terminal hydrolase 4 GRMZM2G128682 7.8 0.97 8.02 pectinesterase 41 GRMZM2G016435 41.18 5.15 7.99 dynein light chain flagellar outer arm
11 GRMZM2G305822 19.35 2.42 7.98 kinase family protein GRMZM2G137435 37.57 4.76 7.89 phospholipase c GRMZM2G125268 40.94 5.2 7.87 mitochondrial aldehyde dehydrogenase lysine-ketoglutarate reductase saccharopine GRMZM2G181362 76.95 9.92 7.76 dehydrogenase bifunctional enzyme GRMZM2G014560 11.16 1.44 7.74 wall-associated kinase 4 GRMZM2G132547 78.97 10.2 7.74 ctp synthase GRMZM2G094990 424 55.13 7.69 beta-expansin 1a precursor GRMZM2G052100 1270.34 165.33 7.68 abscisic stress ripening protein 1 GRMZM2G042347 22.4 2.93 7.65 peroxidase precursor GRMZM2G028926 10.95 1.43 7.63 calcium-dependent protein kinase GRMZM2G144739 5.49 0.72 7.59 exocyst complex subunit sec15-like family protein GRMZM2G325477 116.2 15.33 7.58 pgps d12 GRMZM2G055883 39.05 5.15 7.58 dynamin-related protein expressed GRMZM2G134367 34 4.5 7.55 hypothetical protein LOC100382106 [Zea mays] GRMZM2G134054 11.77 1.56 7.53 purple acid phosphatase isoform b GRMZM2G454449 29.66 3.94 7.53 myb-like dna-binding shaqkyf class family protein GRMZM2G313649 5.66 0.75 7.53 tp53 regulating kinase GRMZM2G134888 36.1 4.82 7.48 amino acid permease GRMZM2G173315 53.72 7.2 7.46 udp-glycosyltransferase-like protein xyloglucan endotransglycosylase hydrolase protein GRMZM2G119783 315.36 42.57 7.41 8 precursor GRMZM2G175576 15.62 2.12 7.37 heavy metal transporter 3 GRMZM5G815358 47.91 6.54 7.33 cold regulated protein 27 GRMZM2G106344 17.55 2.4 7.32 protein disulfide isomerase GRMZM2G154036 28.84 3.95 7.3 neurogenic locus notch protein precursor-like GRMZM2G042789 156.78 21.47 7.3 mpi GRMZM2G361064 300.56 41.22 7.29 expansin os-expa2 GRMZM2G136300 21.52 2.95 7.28 amino acid permease 1
12 GRMZM2G129266 12.39 1.7 7.27 10-deacetylbaccatin iii 10-o-acetyltransferase GRMZM2G334592 14.98 2.08 7.2 s-receptor kinase GRMZM2G336962 108.57 15.09 7.19 bromodomain-containing protein GRMZM2G328500 143.7 20.07 7.16 udp-glucose 6- expressed GRMZM2G154146 15.57 2.18 7.14 nitrate reductase apoenzyme GRMZM2G153208 22.75 3.2 7.12 pr17c precursor GRMZM2G124365 12.41 1.75 7.11 chorismate mutase GRMZM2G000980 4.22 0.59 7.09 splicing factor 1 GRMZM2G180659 153.88 21.96 7.01 amino acid bowman-birk type wound-induced proteinase GRMZM2G011523 735.04 105.19 6.99 inhibitor wip1 precursor GRMZM2G052562 138.18 19.78 6.98 alpha-l-fucosidase 2 precursor GRMZM2G033555 8.18 1.18 6.95 cinnamoyl- reductase GRMZM5G852177 16.61 2.39 6.94 atp binding GRMZM2G063069 32.72 4.72 6.94 protein kinase GRMZM2G081652 36.5 5.29 6.9 cytosolic factor-like protein GRMZM2G026889 10.33 1.5 6.87 transferring glycosyl groups GRMZM2G077655 9.6 1.4 6.85 wall-associated kinase 4 GRMZM2G430849 8.33 1.22 6.83 nac domain transcription expressed GRMZM2G317900 9.2 1.35 6.83 auxin response factor 7a GRMZM2G055999 10.01 1.48 6.77 heavy-metal-associated domain-containing protein GRMZM2G103342 117.1 17.36 6.75 peroxidase 12 GRMZM2G101282 20.2 3 6.74 ubiquitin domain containing 1 GRMZM2G312877 30.39 4.54 6.69 lactoylglutathione lyase GRMZM2G171060 12.63 1.9 6.66 adp-ribosylation factor-like protein GRMZM2G093826 19.68 2.97 6.64 high-affinity potassium transporter pyrophosphate-dependent phosphofructokinase GRMZM2G450163 50.22 7.59 6.62 alpha subunit GRMZM2G409213 14.71 2.23 6.59 mitochondrial glycoprotein family protein
13 GRMZM2G054225 15.27 2.32 6.58 rna polymerase iv second largest subunit GRMZM2G033515 37.91 5.77 6.57 nadp-specific isocitrate dehydrogenase GRMZM2G021621 80.25 12.22 6.57 beta-expansin 1a expressed GRMZM2G087259 63.34 9.76 6.49 carbonic anhydrase GRMZM2G126706 76.18 11.77 6.47 ribonuclease p GRMZM2G348909 6.75 1.05 6.45 cal GRMZM2G049211 44.91 6.97 6.45 vacuolar sorting receptor 1 expressed GRMZM2G091433 12.71 1.97 6.44 repressor protein GRMZM5G858165 14.21 2.21 6.43 cenp-e-like kinetochore protein GRMZM2G165325 34.61 5.38 6.43 hypothetical protein LOC100275463 [Zea mays] GRMZM2G172230 50.54 7.86 6.43 erd1 protein GRMZM2G089010 22.53 3.51 6.43 ethylene receptor GRMZM5G888894 50.75 7.9 6.42 hypothetical protein LOC100384747 [Zea mays] GRMZM2G026980 86.96 13.65 6.37 xyloglucan endotransglycosylase GRMZM2G404965 12.45 1.96 6.37 hexose carrier protein hex6 tpa_exp: homeodomain leucine zipper family iv GRMZM2G118063 24.23 3.81 6.36 protein GRMZM2G078806 23.98 3.79 6.33 transducin wd-40 repeat-containing protein GRMZM2G180328 10.18 1.61 6.32 nac transcription factor GRMZM2G474153 9.96 1.58 6.32 receptor-type protein kinase lrk1 GRMZM2G397261 22.21 3.52 6.31 hypothetical protein LOC100277784 [Zea mays] GRMZM2G109056 31.84 5.06 6.29 lipoxygenase GRMZM2G180699 14.16 2.25 6.29 pollen-specific c2 domain containing protein GRMZM2G044512 11.07 1.76 6.28 f-box domain containing protein GRMZM2G132478 7.04 1.13 6.23 medium chain fatty acid hydroxylase GRMZM2G091226 28.39 4.56 6.23 ww domain containing expressed GRMZM2G109830 11.54 1.86 6.21 protein kinase receptor type precursor GRMZM2G171277 72.38 11.66 6.21 ring-h2 finger protein atl5f GRMZM2G067702 75.34 12.18 6.19 protein phytoclock 1
14 bgl16_orysj ame: full=beta-glucosidase 16 GRMZM5G810727 51.37 8.32 6.18 short=os4bglu16 flags: precursor GRMZM2G434557 50.39 8.16 6.17 vacuolar iron transporter-like protein GRMZM2G059740 12.18 1.99 6.13 protein kinase GRMZM2G126261 45.39 7.42 6.12 peroxidase poc1 GRMZM2G082302 13.26 2.17 6.12 ariadne-like protein ari5 GRMZM2G444801 64.28 10.52 6.11 sulfate transporter GRMZM2G064091 7.35 1.21 6.08 nitrate transporter GRMZM2G099425 5.61 0.92 6.08 calcium-dependent protein hypothetical protein SORBIDRAFT_06g026470 GRMZM2G076844 148.35 24.49 6.06 [Sorghum bicolor] GRMZM2G093276 39.63 6.55 6.05 zinc transporter GRMZM2G304362 23.87 3.95 6.04 ribonucleotide reductase GRMZM2G113191 11.37 1.89 6.03 carbonic anhydrase GRMZM2G098346 121.03 20.08 6.03 alcohol dehydrogenase GRMZM2G149216 17.58 2.92 6.02 histidine amino acid transporter GRMZM5G826838 26.44 4.4 6.01 glutamate decarboxylase GRMZM2G028980 35.6 5.96 5.98 auxin response transcription factor GRMZM2G009045 6.95 1.17 5.96 mitochondrial phosphate transporter calcium-dependent lipid-binding domain- GRMZM2G312838 41.14 6.9 5.96 containing protein GRMZM2G118800 44.11 7.43 5.94 aldehyde dimeric nadp-preferring GRMZM2G074262 30.13 5.09 5.92 lstk-1-like kinase GRMZM2G060507 65.74 11.11 5.92 homeobox protein expressed GRMZM2G106622 13.99 2.37 5.89 aba-responsive protein GRMZM2G056513 16.85 2.87 5.88 hypothetical protein LOC100275283 [Zea mays] GRMZM2G080045 11.58 1.97 5.87 ammonium transporter GRMZM2G015983 31.39 5.36 5.86 xyloglucan fucosyltransferase GRMZM2G101958 1127.47 192.54 5.86 phospholipid transfer protein 1 GRMZM5G803404 24.93 4.26 5.85 white-brown-complex abc transporter family
15 GRMZM2G083546 9.75 1.68 5.82 zinc finger protein glo3-like GRMZM2G151169 6.81 1.17 5.8 2-oxoglutarate e1 component GRMZM2G104907 15.81 2.73 5.79 vesicle-associated membrane GRMZM2G061702 18.7 3.24 5.78 cyclase family protein GRMZM2G168502 11.65 2.02 5.77 nodulin 21 family expressed GRMZM2G160506 16.92 2.94 5.76 gdp-mannose pyrophosphorylase GRMZM2G069694 104.34 18.14 5.75 plant-specific domain tigr01615 family protein GRMZM2G023436 54.04 9.4 5.75 gtp-binding protein GRMZM2G329002 14.47 2.52 5.73 pectinacetylesterase precursor GRMZM2G123104 7.04 1.23 5.73 Os01g0347300 [Oryza sativa Japonica Group] GRMZM2G109405 20.13 3.53 5.71 purple acid phosphatase 29 GRMZM2G053720 129.84 22.76 5.71 proline oxidase GRMZM2G142409 59.41 10.42 5.7 24 kda seed maturation protein GRMZM2G059693 19.38 3.42 5.66 lipase class 3-like GRMZM2G020940 531.43 93.93 5.66 r40c1 protein - rice GRMZM2G051541 38.99 6.9 5.65 phloem protein 2-b2 GRMZM2G071343 43.59 7.77 5.61 polyamine oxidase GRMZM2G479110 27.9 4.97 5.61 response regulator 9 GRMZM2G155111 23.62 4.22 5.6 protein iq-domain 32 GRMZM2G130943 12.15 2.17 5.6 protein phosphatase xyloglucan endotransglucosylase hydrolase protein GRMZM2G113761 22.46 4.03 5.57 32 precursor GRMZM2G099239 200.58 36.03 5.57 dna binding protein GRMZM2G026470 137.5 24.77 5.55 soluble inorganic pyrophosphatase GRMZM2G321839 35.69 6.44 5.55 peroxidase 1 precursor GRMZM2G467370 36.21 6.54 5.54 unknown [Zea mays] GRMZM2G010960 7.01 1.27 5.54 pollen-specific protein GRMZM2G138624 13.58 2.46 5.53 nicotinamidase 1 AC210173.4_FG005 10.1 1.83 5.52 ferulate-5-hydroxylase
16 vps51 vps67 family (components of vesicular GRMZM2G132991 10.91 1.98 5.52 transport) protein GRMZM2G419239 27.14 4.92 5.51 transcription factor myb42 GRMZM2G119116 23.62 4.29 5.51 desacetoxyvindoline 4- GRMZM2G059497 25.63 4.68 5.47 serine threonine-specific protein kinase npk15-like endoplasmic reticulum-type calcium-transporting GRMZM2G141704 10.62 1.94 5.47 atpase 4 GRMZM2G428216 46.65 8.53 5.47 hypothetical protein LOC100276732 [Zea mays] GRMZM2G025536 9.7 1.77 5.47 fe2+ dioxygenase-like GRMZM2G477325 299.93 54.93 5.46 low temprature induced-like protein GRMZM2G152258 30.4 5.6 5.42 tropinone reductase 2 GRMZM2G018006 49.95 9.23 5.41 1-aminocyclopropane-1-carboxylate synthase GRMZM2G088689 33.73 6.26 5.39 2-oxoisovalerate dehydrogenase e1 alpha subunit GRMZM2G366659 20.29 3.77 5.38 trehalose 6-phosphate synthase GRMZM2G056407 40.52 7.54 5.38 transcription factor myb94 GRMZM2G122853 21.53 4.02 5.36 peroxidase 1 GRMZM2G168744 12.35 2.31 5.36 bifunctional nuclease bfn1 GRMZM2G361256 19.75 3.69 5.35 mrp-like abc transporter GRMZM2G066923 55.52 10.39 5.34 equilibrative nucleoside transporter GRMZM2G121996 13.7 2.57 5.34 methyltransferase pmt2 GRMZM2G113726 19.01 3.56 5.34 wd-40 repeat family protein GRMZM2G066870 66.62 12.49 5.33 low temprature induced-like protein AC194974.3_FG005 6.11 1.15 5.32 receptor-type protein kinase lrk1 GRMZM2G163578 168.72 31.72 5.32 sigma factor sigb regulation protein GRMZM2G094541 17.57 3.31 5.31 serine threonine protein kinase GRMZM2G129189 146.81 27.65 5.31 class iv chitinase GRMZM2G059637 7.88 1.48 5.31 glycerol-3-phosphate acyltransferase 5 GRMZM2G151268 15.81 2.99 5.29 tonneau 1b GRMZM2G156861 933.81 176.59 5.29 lipoxygenase
17 GRMZM2G337048 25.31 4.81 5.26 udp-glycosyltransferase-like protein GRMZM2G456217 48.1 9.14 5.26 cysteine proteinase GRMZM2G113613 86.82 16.51 5.26 eps15 homology domain 1 protein GRMZM2G090842 39.03 7.46 5.23 act domain containing expressed GRMZM2G400470 8.32 1.59 5.23 map kinase kinase GRMZM2G067789 44.12 8.5 5.19 d-xylose-proton symporter-like 2 GRMZM2G374969 47.43 9.29 5.11 wd-repeat protein GRMZM2G110567 135.26 26.51 5.1 protein binding GRMZM2G078959 6.06 1.19 5.1 tpx2 (targeting protein for xklp2) family protein GRMZM2G118821 58.34 11.47 5.09 zip transporter t7 bacteriophage-type single subunit rna GRMZM2G381395 36.77 7.23 5.09 polymerase GRMZM2G450233 554.52 109.08 5.08 peroxidase GRMZM2G151967 27.32 5.38 5.08 multiple inositol polyphosphate phosphatase a1 GRMZM2G445602 593.61 117.19 5.07 fiddlehead-like protein GRMZM2G166459 9.7 1.92 5.06 ripening regulated protein GRMZM2G137582 8.78 1.74 5.04 ein3-binding f-box protein 1 GRMZM2G045809 16.45 3.26 5.04 germin protein type 1 GRMZM5G862540 104.39 20.73 5.03 udp-glucose 6- expressed GRMZM2G054803 31.16 6.19 5.03 undecaprenyl pyrophosphate synthetase GRMZM2G146173 27.7 5.51 5.03 zinc finger protein hangover GRMZM2G126062 6.41 1.28 5.02 nad h-dependent oxidoreductase GRMZM2G113967 111.28 22.18 5.02 cbl-interacting protein kinase GRMZM2G011513 24.38 4.87 5.01 c2 domain-containing protein GRMZM2G142757 759.46 151.75 5 clathrin light chain protein GRMZM2G052650 17.62 3.55 4.96 atp binding GRMZM2G164974 170.54 34.5 4.94 at1g68530 t26j14_10 GRMZM2G152751 33.88 6.86 4.94 hypothetical protein LOC100191329 [Zea mays] GRMZM5G863420 19.71 3.99 4.93 wrky transcription factor
18 GRMZM2G116646 31.43 6.37 4.93 axial regulator yabby2 GRMZM2G088053 12.1 2.46 4.92 nodulin 21 family expressed GRMZM2G162531 46.26 9.44 4.9 strubbelig-receptor family 8 protein GRMZM2G125557 80.73 16.48 4.9 dormancy auxin associated-like protein AC189879.3_FG003 48.91 10.03 4.88 eukaryotic translation initiation factor GRMZM2G004548 68 13.98 4.87 zinc-binding protein GRMZM2G361593 17.86 3.67 4.87 myo-inositol kinase GRMZM2G035821 29.28 6.04 4.85 s-ribonuclease binding protein GRMZM2G358830 8.63 1.78 4.85 receptor protein kinase perk1 GRMZM2G450512 266.49 55.02 4.84 sulfur-rich thionin-like protein GRMZM2G074850 22.68 4.68 4.84 gdp-mannose dehydratase 2 GRMZM2G089854 25.06 5.19 4.82 auxin-independent growth promoter GRMZM2G142718 13.47 2.79 4.82 dof zinc finger protein GRMZM2G022642 14.55 3.03 4.81 amino acid transporter GRMZM2G176394 108.36 22.56 4.8 leucine-rich receptor-like protein kinase GRMZM2G121649 105.66 22.03 4.8 microtubule-associated protein 65-7 GRMZM2G429714 60.66 12.7 4.78 receptor protein GRMZM2G134708 20.7 4.34 4.77 monodehydroascorbate reductase GRMZM2G083526 167.03 35.08 4.76 gl1 protein GRMZM2G165357 123.88 26.03 4.76 udp-xyl synthase 6 GRMZM2G101687 8.52 1.79 4.75 serine threonine-protein kinase nak GRMZM2G128880 16.11 3.4 4.74 nicotinamidase 1 GRMZM2G176723 46.42 9.82 4.73 gcn5-related n-acetyltransferase-like protein GRMZM2G305264 27.26 5.8 4.7 protein binding protein GRMZM2G384439 92.81 19.76 4.7 somatic embryogenesis receptor-like kinase 1 GRMZM2G036448 205.71 43.83 4.69 amino acid glycosyl hydrolase family 3 c terminal domain GRMZM2G035503 18.23 3.88 4.69 containing expressed GRMZM2G315072 35.06 7.48 4.69 ankyrin repeat-rich protein
19 GRMZM2G156026 11.72 2.5 4.69 hydroquinone glucosyltransferase GRMZM2G418232 21.58 4.61 4.68 ubiquitin carboxyl-terminal hydrolase 12 GRMZM2G465226 36.97 7.91 4.67 pathogenesis-related protein class i GRMZM5G853854 37.66 8.07 4.67 glycolate oxidase GRMZM5G883759 25.56 5.49 4.66 extensin-like cell wall protein GRMZM2G312661 40.07 8.64 4.64 ef hand family expressed GRMZM2G012160 20.96 4.53 4.63 cysteine proteinase inhibitor GRMZM2G123667 12.11 2.62 4.62 5 protein GRMZM2G170760 15.48 3.36 4.6 clathrin assembly protein GRMZM2G116584 56.61 12.34 4.59 calcineurin b-like protein 10 GRMZM2G126010 16.24 3.54 4.58 actin- expressed GRMZM2G099049 113.86 24.87 4.58 auxin-repressed protein GRMZM2G003794 12.11 2.65 4.57 endo- -beta-xylanase GRMZM2G127072 91.82 20.1 4.57 expansin-like 3 precursor lag1 and cln8 lipid-sensing domain containing GRMZM2G020148 84.39 18.48 4.57 protein GRMZM2G108084 17.95 3.94 4.56 protein GRMZM2G351330 12.75 2.8 4.55 homeodomain leucine zipper protein GRMZM2G010868 45.43 9.98 4.55 lipid-transfer protein GRMZM2G077942 38.01 8.36 4.55 actin depolymerizing factor 5 GRMZM2G066441 14.88 3.28 4.54 cytochrome p450 homeodomain-leucine zipper transcription factor GRMZM2G041462 23.1 5.09 4.54 ipi-1 AC233893.1_FG003 84.36 18.6 4.54 at1g68530 t26j14_10 GRMZM2G156310 28.02 6.19 4.53 gibberellin receptor gid1l2 GRMZM2G127141 28.25 6.26 4.52 big map kinase GRMZM2G014994 11.84 2.62 4.51 gpi-anchored protein GRMZM2G148200 19.74 4.38 4.51 vacuolar proton-inorganic pyrophosphatase GRMZM2G056247 13.42 2.98 4.5 cytochrome p450
20 GRMZM2G126749 21.12 4.71 4.49 calmodulin-binding protein GRMZM2G130558 76.45 17.04 4.49 plastidic phosphate translocator-like protein1 GRMZM2G032028 24.95 5.56 4.49 hypothetical protein LOC100191857 [Zea mays] GRMZM2G094589 87.35 19.47 4.49 harpin-induced protein 1 containing expressed GRMZM2G135091 11.34 2.54 4.47 phosphoenolpyruvate carboxylase kinase GRMZM2G074759 21.7 4.86 4.47 amp dependent GRMZM2G154678 11.36 2.54 4.47 endo- -beta- GRMZM2G144109 46.31 10.37 4.47 protein phosphatase GRMZM2G013236 88.18 19.76 4.46 cbl-interacting protein kinase 23 GRMZM2G011078 19.8 4.44 4.46 sc3 protein GRMZM2G104176 38.3 8.6 4.45 iaa16 - auxin-responsive aux iaa family member GRMZM2G061723 20.82 4.68 4.45 calmodulin-binding protein GRMZM2G147619 7.56 1.7 4.45 histone-lysine n-methyltransferase ashh1 GRMZM2G075562 80.47 18.19 4.42 zinc finger protein constans-like 9 GRMZM2G058310 21.13 4.78 4.42 beta-amylase GRMZM2G441541 114.23 25.9 4.41 nadph oxidase GRMZM2G098239 256.1 58.17 4.4 hxxxd-type acyl-transferase-like protein GRMZM2G134264 643.19 146.58 4.39 pore-forming toxin-like protein hfr-2 18s pre-ribosomal assembly protein gar2-related GRMZM2G131727 9.95 2.27 4.39 protein GRMZM2G007249 15.01 3.43 4.38 1-aminocyclopropane-1-carboxylate oxidase GRMZM2G345544 16.32 3.73 4.37 copine i AC191097.3_FG007 52.94 12.12 4.37 cytochrome b561 GRMZM2G157564 79.18 18.14 4.36 white-brown-complex abc transporter family GRMZM2G381473 27.33 6.27 4.36 udp-glucuronic acid decarboxylase GRMZM2G337330 400.26 91.84 4.36 hypothetical protein LOC100275514 [Zea mays] GRMZM2G451716 94.86 21.81 4.35 mei2-like protein GRMZM2G001289 48.38 11.14 4.34 roc1(homeobox protein) GRMZM2G040452 162.5 37.4 4.34 protein phosphatase-
21 GRMZM2G082642 8.12 1.87 4.34 spla ryanodine receptor domain-containing protein hypothetical protein SORBIDRAFT_03g008020 GRMZM2G410916 39.78 9.16 4.34 [Sorghum bicolor] transmembrane emp24 domain-containing protein GRMZM2G136058 86.3 19.96 4.32 10 precursor GRMZM2G135739 63.76 14.75 4.32 monosaccharide transporter 3 GRMZM2G060760 37.4 8.65 4.32 alpha beta fold family expressed GRMZM2G060148 21.96 5.1 4.31 hypothetical protein [Zea mays] GRMZM2G167637 25.13 5.84 4.3 pectin methylesterase GRMZM2G398506 12.83 2.98 4.3 wrky transcription GRMZM2G004349 11.98 2.79 4.3 seed maturation protein GRMZM2G002131 12.52 2.91 4.29 heat shock transcription factor hsf4 GRMZM2G176780 14.88 3.47 4.29 carbohydrate transporter sugar porter transporter GRMZM2G152461 11.1 2.6 4.27 ring finger protein 5 GRMZM5G863596 34.88 8.18 4.26 plastid alpha-amylase GRMZM2G031317 28.41 6.68 4.26 phytosulfokines 2 precursor GRMZM2G094712 20.02 4.71 4.25 aspartate aminotransferase GRMZM2G174830 14.4 3.39 4.24 kelch repeat-containing protein GRMZM2G012306 21.52 5.07 4.24 peptide transporter ptr2 GRMZM2G040030 58.68 13.85 4.24 cold acclimation protein cor413-pm1 GRMZM2G448523 117.38 27.71 4.24 aig1 family expressed dnaj heat shock n-terminal domain-containing GRMZM2G168397 26.35 6.24 4.22 protein GRMZM2G451965 26.85 6.36 4.22 endo-beta- -glucanase precursor GRMZM2G004748 17.76 4.22 4.21 multidrug pheromone mdr abc transporter family GRMZM2G152975 30.52 7.25 4.21 alcohol dehydrogenase GRMZM2G452142 15.37 3.65 4.21 phytosulfokine receptor GRMZM2G147046 29.99 7.13 4.21 coated vesicle membrane GRMZM2G161809 9 2.14 4.2 transparent testa 12 protein GRMZM2G076778 26.54 6.31 4.2 hypothetical protein LOC100276620 [Zea mays]
22 GRMZM2G075496 14.96 3.56 4.2 osbp(oxysterol binding protein)-related protein 4c GRMZM2G013391 23.79 5.69 4.18 wrky transcription factor GRMZM5G870077 192.32 46.01 4.18 f-box family protein GRMZM2G071871 24.31 5.82 4.18 aspartyl transcarbamylase GRMZM2G098637 17.27 4.15 4.16 e3 ubiquitin-protein ligase rhf2a GRMZM2G143782 18.52 4.46 4.15 speckle-type poz protein GRMZM2G073725 139.12 33.51 4.15 reversibly glycosylated polypeptide GRMZM2G010054 5.55 1.34 4.15 coatomer alpha GRMZM2G481730 12.32 2.97 4.15 rna-dependent rna polymerase GRMZM2G128564 17.38 4.2 4.14 hxxxd-type acyl-transferase-like protein GRMZM2G459755 21.25 5.14 4.14 aim1 protein GRMZM2G151997 15.4 3.75 4.1 hypothetical protein LOC100277656 [Zea mays] GRMZM2G118737 17.44 4.26 4.09 neutral alkaline invertase GRMZM2G038281 18.17 4.44 4.09 beta-galactosidase 10 GRMZM2G007195 32.05 7.85 4.08 udp-glucuronic acid decarboxylase GRMZM2G159500 36.53 8.96 4.07 nac domain containing protein 87 hypothetical protein SORBIDRAFT_04g002640 GRMZM2G017229 14.14 3.47 4.07 [Sorghum bicolor] GRMZM2G026065 10.05 2.47 4.07 protein kinase g11a GRMZM2G115773 11.17 2.76 4.05 xylem serine proteinase 1 GRMZM2G054559 34.13 8.43 4.05 phospholipase d GRMZM2G050984 8.82 2.18 4.05 xanthine dehydrogenase GRMZM2G028521 258.6 63.92 4.05 transmembrane expressed GRMZM2G169967 45.11 11.15 4.04 beta-expansin 3 GRMZM2G059893 7.67 1.9 4.04 kinase family protein GRMZM2G108537 37.54 9.29 4.04 nodulin 21 family expressed GRMZM2G359322 30.98 7.68 4.04 elf4-like protein GRMZM2G144782 152.08 37.71 4.03 chy zinc finger family expressed GRMZM2G000741 40.63 10.08 4.03 mitochondrial substrate carrier family protein
23 GRMZM2G166218 25.43 6.33 4.01 pollen ole e 1 allergen and extensin family protein GRMZM2G177812 148.03 36.93 4.01 atp-binding cassette GRMZM2G010823 25.06 6.25 4.01 fumarate hydratase 2 GRMZM2G166176 15.82 3.96 4 er glycerol-phosphate acyltransferase GRMZM2G114192 30.72 7.69 3.99 rac gtpase activating protein 2 GRMZM5G828396 60.88 15.28 3.99 bhlh transcription factor GRMZM2G083725 260.74 65.48 3.98 lipid transfer protein GRMZM2G144668 32.33 8.13 3.98 desacetoxyvindoline 4- GRMZM5G876616 178.31 44.85 3.98 lurp-one-related 15 protein GRMZM2G098397 11.97 3.02 3.96 heparanase-like protein 2 precursor GRMZM2G020761 40.13 10.12 3.96 cytochrome p450 GRMZM2G047949 24.74 6.24 3.96 superkiller viralicidic activity 2-like 2-like GRMZM2G008226 22.37 5.66 3.95 trehalose 6-phosphate synthase GRMZM2G029242 28.5 7.23 3.94 serine threonine-protein kinase receptor precursor GRMZM2G411668 71.54 18.15 3.94 s-locus lectin protein kinase family protein GRMZM2G312521 170.38 43.25 3.94 trehalose-6-phosphate synthase hypothetical protein Z273B07_Z409L08.26 [Zea GRMZM2G310453 43.01 10.92 3.94 mays] GRMZM2G074914 36.5 9.29 3.93 catalytic hydrolase GRMZM2G066191 32.81 8.36 3.93 beta-4 tubulin GRMZM2G173124 24.76 6.32 3.92 ccch-type zinc finger protein GRMZM2G352618 58.51 14.94 3.92 hypothetical protein LOC100277700 [Zea mays] rossmann-fold nad -binding domain-containing GRMZM2G135277 23.39 5.98 3.91 protein GRMZM2G180988 20.06 5.13 3.91 villin 2 GRMZM5G885329 205.58 52.57 3.91 nac domain ipr003441 GRMZM2G080041 24.28 6.21 3.91 flagellin-sensing 2-like protein GRMZM2G052630 10.76 2.76 3.9 isovaleryl- dehydrogenase GRMZM2G089783 38.22 9.82 3.89 prenylated rab acceptor family protein
24 GRMZM2G432796 12.77 3.29 3.88 protein kinase domain-containing protein GRMZM2G443525 65.95 16.99 3.88 leucine rich repeat family expressed GRMZM2G161411 21.13 5.45 3.88 dna-binding protein GRMZM2G048129 60.59 15.67 3.87 mtn19-like protein GRMZM2G009103 19.78 5.12 3.86 nodulin-like protein GRMZM2G020146 9.62 2.49 3.86 serine carboxypeptidase iii precursor GRMZM2G474119 40.73 10.56 3.86 hypothetical protein LOC100501037 [Zea mays] GRMZM2G009479 55.93 14.51 3.86 lipoxygenase GRMZM5G802115 10.79 2.8 3.85 kinase family protein GRMZM2G047476 198.23 51.56 3.84 vacuolar atp synthase 16 kda proteolipid subunit GRMZM2G388585 18.31 4.76 3.84 related GRMZM2G065696 15.43 4.02 3.84 plac8-like protein GRMZM2G076810 29.99 7.81 3.84 hypothetical protein [Zea mays] GRMZM2G046743 17.84 4.65 3.84 amino acid GRMZM2G434696 19.63 5.12 3.84 nucleotide binding GRMZM2G179090 27.56 7.19 3.84 ras-related protein GRMZM2G431708 15.04 3.93 3.83 ketose-bisphosphate aldolase class-ii-like protein GRMZM2G333875 23.76 6.22 3.82 receptor protein kinase-like GRMZM2G136580 48.75 12.82 3.8 glycosyltransferase family 61 protein GRMZM2G135108 32.22 8.48 3.8 peroxidase GRMZM2G147245 12.21 3.21 3.8 trans-cinnamate 4-monooxygenase GRMZM2G176085 71.49 18.84 3.79 peroxidase 73 GRMZM2G101523 6.82 1.8 3.79 phosphoinositide binding protein GRMZM2G051677 172.4 45.45 3.79 fructokinase-2 GRMZM2G027333 19.66 5.2 3.78 c2h2l domain class transcription factor GRMZM2G307252 41.75 11.04 3.78 sec61 transport protein GRMZM2G363813 8.86 2.34 3.78 hipl1 protein GRMZM2G126865 14.13 3.74 3.78 endosomal p24a protein GRMZM2G316474 29.15 7.73 3.77 serine threonine-protein kinase
25 GRMZM2G377131 29 7.7 3.77 senescence-associated protein GRMZM2G332809 7.33 1.95 3.76 beta-tubulin GRMZM2G176455 32.26 8.58 3.76 rex1 dna repair family protein GRMZM2G109560 62.6 16.66 3.76 lon protease GRMZM2G055180 32.47 8.65 3.76 ethylene-responsive transcription factor 2 GRMZM5G854613 65.65 17.5 3.75 aim1 protein GRMZM2G104538 40.94 10.91 3.75 phytol kinase GRMZM2G157168 36.99 9.87 3.75 amino acid expressed GRMZM2G092101 12.4 3.32 3.74 kelch repeat-containing f-box family expressed GRMZM2G053916 41.46 11.1 3.74 electron-transfer- alpha polypeptide GRMZM2G032218 7.86 2.11 3.73 abc transporter c family member 9 GRMZM2G017557 18.87 5.06 3.73 o-methyltransferase zrp4 GRMZM2G113512 20.84 5.59 3.73 protein kinase domain-containing protein GRMZM2G147726 97.32 26.11 3.73 adp-ribosylation factor 3 GRMZM2G427618 19.73 5.3 3.73 gibberellin 2-oxidase GRMZM2G067511 31.48 8.47 3.72 calmodulin GRMZM2G137558 6.33 1.7 3.71 phosphatidylinositol 3- and 4-kinase family-like GRMZM2G143494 74.51 20.11 3.7 strictosidine synthase GRMZM2G127948 219.11 59.22 3.7 caffeoyl- 3-o-methyltransferase 1 GRMZM2G151763 12.09 3.27 3.69 wrky dna-binding protein 28 GRMZM2G391794 36.73 9.94 3.69 receptor protein kinase clavata1 GRMZM2G346133 124.73 33.77 3.69 dynein light chain flagellar outer arm GRMZM2G122767 16.24 4.41 3.68 cytosolic delta subunit GRMZM2G152179 14.85 4.04 3.68 steroid sulfotransferase 3 GRMZM2G145715 53.48 14.59 3.67 6-phosphogluconate dehydrogenase GRMZM2G127123 50.21 13.71 3.66 beta-galactosidase expressed GRMZM2G007404 31.99 8.74 3.66 udp-glucuronic acid decarboxylase calcium-dependent lipid-binding domain- GRMZM2G082199 40.94 11.2 3.65 containing protein
26 GRMZM2G527891 9.91 2.71 3.65 trehalose 6-phosphate synthase GRMZM2G308860 32.32 8.86 3.65 abc transporter GRMZM2G078440 268.36 73.55 3.65 nucleic acid binding GRMZM2G061321 13.68 3.76 3.64 anthocyanidin 3-o-glucosyltransferase GRMZM5G831142 11.81 3.25 3.64 transducin wd40 domain-containing protein GRMZM2G116079 88.58 24.35 3.64 rna-binding protein cabeza GRMZM2G035405 33.27 9.17 3.63 auxin response factor GRMZM2G126732 18.83 5.2 3.62 1-aminocyclopropane-1-carboxylate oxidase GRMZM2G073860 51.4 14.24 3.61 purple acid phosphatase GRMZM2G111017 15.22 4.22 3.6 adp atp translocase GRMZM2G082191 16.77 4.66 3.6 leucine-rich repeat receptor-like protein kinase GRMZM2G092468 25.34 7.04 3.6 charged multivesicular body protein 5 GRMZM2G007206 58.22 16.21 3.59 tetraspanin family protein GRMZM2G027098 90.26 25.24 3.58 tonoplast intrinsic protein GRMZM2G019619 81.3 22.78 3.57 glucan endo- -beta-glucosidase 7 precursor GRMZM2G414727 53.2 14.92 3.57 saur-like auxin-responsive protein GRMZM2G013750 21.36 5.99 3.56 26s proteasome regulatory subunit n5 GRMZM5G822591 10.91 3.07 3.56 heavy chain GRMZM2G169994 21.13 5.94 3.56 ring domain containing protein two-component system sensor histidine kinase GRMZM2G060485 15.21 4.29 3.55 response GRMZM2G305856 97.06 27.38 3.54 myb transcription factor GRMZM2G107565 9.9 2.79 3.54 protein phosphatase 2c 52 GRMZM2G044629 68.81 19.42 3.54 udp-n-acetylglucosamine pyrophosphorylase GRMZM2G066290 13.1 3.71 3.53 pyruvate kinase GRMZM2G101117 60.01 17.01 3.53 anter-specific proline-rich protein apg GRMZM2G001088 15.43 4.37 3.53 sas10 utp3 c1d family GRMZM5G822137 42.02 11.95 3.52 heavy metal-associated domain containing protein GRMZM2G050467 47.08 13.42 3.51 proteasome subunit alpha type 7
27 GRMZM5G835039 46.07 13.16 3.5 hypothetical protein LOC100275470 [Zea mays] GRMZM2G044963 24.49 7.01 3.49 calmodulin GRMZM2G150912 36.21 10.37 3.49 meiotically up-regulated gene 71 GRMZM2G131982 62.7 17.97 3.49 salt tolerance-like protein GRMZM2G314898 80.13 23.01 3.48 3-n-debenzoyl-2-deoxytaxol n-benzoyltransferase GRMZM2G051103 27.95 8.03 3.48 cbl-interacting serine threonine-protein kinase 9 GRMZM2G044724 43.38 12.47 3.48 leucine rich repeat family expressed GRMZM2G142544 46.88 13.49 3.48 protein kinase GRMZM2G121022 43.48 12.51 3.48 signal recognition particle 68 kda GRMZM2G047093 18.56 5.35 3.47 tetratricopeptide repeat-containing protein GRMZM2G009144 25.02 7.22 3.46 hypothetical protein LOC100381587 [Zea mays] GRMZM2G099481 16.31 4.71 3.46 lipin-like protein GRMZM2G022403 61.62 17.83 3.46 membrane protein GRMZM2G061745 109.53 31.84 3.44 26s proteasome regulatory subunit n8 GRMZM2G148460 17.29 5.04 3.43 esterase precursor GRMZM2G081582 44.54 13 3.43 phenylalanine ammonia-lyase dolichyl-diphosphooligosaccharide--protein GRMZM2G462325 28.77 8.41 3.42 glycosyltransferase 48 kdasubunit precursor hypothetical protein SORBIDRAFT_06g026470 GRMZM2G018027 18 5.26 3.42 [Sorghum bicolor] GRMZM2G173647 13.39 3.92 3.42 leucine-rich repeat-containing protein GRMZM2G326235 332.82 97.49 3.41 hypothetical protein [Zea mays] GRMZM2G064255 267.74 78.43 3.41 glutathione -s-transferase GRMZM2G118265 14.58 4.29 3.4 copine i-like GRMZM2G035552 27.74 8.16 3.4 act domain containing expressed GRMZM2G090087 57 16.79 3.4 aldehyde dehydrogenase GRMZM2G035741 453.8 133.67 3.39 91a protein GRMZM2G004475 18.05 5.33 3.39 wd-40 repeat protein GRMZM2G115000 20.12 5.95 3.38 u-box domain-containing protein 7
28 GRMZM2G081915 40.4 11.95 3.38 serine threonine-protein kinase glycoprotein membrane precursor gpi-anchored GRMZM2G322618 44.68 13.24 3.37 protein GRMZM2G114873 81.47 24.16 3.37 dna binding GRMZM2G151087 51.02 15.16 3.37 3-hydroxyacyl- dehydratase pasticcino 2 GRMZM2G143354 14.14 4.21 3.36 coatomer subunit delta haloacid dehalogenase-like hydrolase domain- GRMZM2G179432 760.94 227.28 3.35 containing protein GRMZM2G012501 12.89 3.85 3.35 coatomer subunit gamma GRMZM2G104626 61.28 18.34 3.34 fatty acid elongase 3-ketoacyl- synthase 1 AC209898.2_FG004 31.22 9.35 3.34 gcip-interacting family GRMZM2G121565 26.42 7.92 3.34 protein kinase GRMZM2G179985 20.07 6.02 3.34 lung seven transmembrane receptor 1 GRMZM2G141473 35.71 10.71 3.33 aldehyde oxidase GRMZM2G409722 37.52 11.26 3.33 syringolide-induced protein 14-1-1 GRMZM2G069923 104.36 31.35 3.33 ring-h2 finger protein atl2b GRMZM2G180372 26.7 8.04 3.32 hypothetical protein [Zea mays] GRMZM2G412846 90.93 27.46 3.31 phosphatidylinositol-4-phosphate 5-kinase GRMZM2G035584 149.51 45.19 3.31 anthranilate n-benzoyltransferase protein 1 GRMZM2G320506 21.05 6.37 3.31 calcium-dependent protein kinase GRMZM2G054807 21.34 6.48 3.29 avr9 cf-9 rapidly elicited protein 137 GRMZM2G430600 155.62 47.34 3.29 calmodulin GRMZM2G341934 24.39 7.42 3.29 peroxidase family expressed GRMZM2G055698 71.92 21.93 3.28 membrane protein GRMZM2G176206 25.01 7.66 3.26 clavata1 receptor kinase GRMZM2G010460 55.46 17.06 3.25 ubiquitin-conjugating enzyme -like brassinosteroid insensitive 1-associated receptor GRMZM2G125308 24.38 7.5 3.25 kinase 1 precursor GRMZM2G431243 31.71 9.76 3.25 iq calmodulin-binding motif family expressed GRMZM2G145827 27.45 8.45 3.25 dynamin-related protein 1c
29 GRMZM2G162413 29.67 9.18 3.23 auxin-responsive protein GRMZM2G031001 35.93 11.12 3.23 nac domain containing protein 87 GRMZM2G110541 25.05 7.78 3.22 zinc finger protein AC199765.4_FG008 20.82 6.47 3.22 membrane-anchored endo- -beta-glucanase GRMZM2G006937 33.07 10.32 3.21 cycloartenol synthase GRMZM2G118917 18.07 5.64 3.2 exosome complex exonuclease rrp6 GRMZM2G127798 96.33 30.15 3.2 6-phosphogluconate dehydrogenase GRMZM2G131280 106.75 33.55 3.18 platz transcription factor GRMZM2G406672 20.15 6.34 3.18 malate oxidoreductase (malic enzyme) short chain dehydrogenase reductase family GRMZM2G128577 72.74 22.9 3.18 expressed GRMZM2G442658 48.08 15.14 3.17 alcohol dehydrogenase 1 GRMZM2G064541 6.88 2.17 3.16 nac domain ipr003441 GRMZM2G019986 33.44 10.59 3.16 udp-n-acetylglucosamine pyrophosphorylase GRMZM2G071087 217 68.81 3.15 protein phosphatase 2c containing expressed GRMZM2G079471 21.38 6.78 3.15 zinc binding GRMZM2G015735 21.31 6.77 3.15 hypothetical protein LOC100304181 [Zea mays] GRMZM2G314396 12.8 4.07 3.14 calcium-dependent protein kinase GRMZM2G152908 112.9 35.92 3.14 sucrose synthase GRMZM2G026147 208.66 66.41 3.14 expansin-like 3 precursor GRMZM2G153333 15.53 4.94 3.14 gras family transcription factor GRMZM5G838098 62.87 20.02 3.14 transcription factor GRMZM2G092129 45.7 14.57 3.14 integral membrane single c2 domain protein GRMZM2G317770 31.85 10.18 3.13 receptor-like kinase GRMZM2G033641 37.07 11.86 3.13 cellular retinaldehyde-binding triple function brassinosteroid insensitive 1-associated receptor GRMZM5G867798 17.87 5.72 3.12 kinase 1 expressed GRMZM2G108138 50.38 16.13 3.12 acyl binding expressed GRMZM2G142063 183.68 58.84 3.12 inositol transporter 2
30 GRMZM2G358059 57.19 18.32 3.12 calreticulin precursor GRMZM2G379540 86.27 27.66 3.12 membrane protein GRMZM2G173680 13.6 4.37 3.11 wrky dna binding domain containing expressed GRMZM2G062156 52.5 16.9 3.11 sorbitol transporter GRMZM2G463462 54.79 17.75 3.09 vacuolar iron transporter-like protein GRMZM2G114162 29.14 9.49 3.07 phosphatidylinositol 4-kinase alpha GRMZM2G424832 68.97 22.57 3.06 cellulose synthase GRMZM2G178025 11.49 3.76 3.05 membrane-anchored endo- -beta-glucanase GRMZM2G105415 47.51 15.58 3.05 protein kinase family protein GRMZM2G053531 22.09 7.3 3.02 wound responsive protein GRMZM2G452896 16.46 5.45 3.02 serine carboxypeptidase s28 family protein 2-oxoisovalerate dehydrogenase e2 component GRMZM2G033644 44.71 14.82 3.02 (dihydrolipoyl transacylase) GRMZM2G143205 30.95 10.27 3.01 calmodulin-binding transcription activator 1 MT, mutant; WT, wild type.
TableS3 Genes completely suppressed in heterozygous Rg1 leaves compared to the wild type siblings
FPKM value Gene ID Annotation of WT GRMZM2G140107 865577 sucrose phosphate synthase GRMZM2G127635 13.64 hypothetical protein LOC100192806 [Zea mays] GRMZM2G048904 11.55 alpha-l-fucosidase 2 GRMZM2G158117 9.51 sant myb protein GRMZM2G443265 6.9 organic anion transporter
31 TableS4 Genes down-regulated more than 3-fold change in heterozygous Rg1 leaves compared to the wild type siblings
FPKM value FPKM value Fold change Gene ID Annotation of MT of WT WT/MT GRMZM2G093950 24.31 712851 29325.78 polynucleotide adenylyltransferase-like protein GRMZM2G049378 7.39 181.23 24.54 sant myb protein GRMZM2G082032 8.19 166.3 20.31 isoaspartyl peptidase l-asparaginase 2 subunit beta GRMZM2G051101 1.18 23.63 19.96 ubiquitin ligase GRMZM2G045154 0.52 9.45 18.28 pentatricopeptide repeat-containing protein GRMZM5G891187 1.18 17.76 15.06 nodulin-like protein GRMZM2G108597 1.73 20.41 11.8 transmembrane amino acid transporter family protein GRMZM2G114748 8.27 93.27 11.28 regulator of chromosome condensation repeat-containing protein GRMZM2G038519 20.21 209.42 10.36 chlorophyll a-b binding protein GRMZM2G138840 3.19 28.23 8.85 transducin wd40 domain-containing protein GRMZM2G115817 8.94 78.45 8.77 circadian clock coupling factor zgt GRMZM2G524937 2.18 18.82 8.63 swib domain-containing protein GRMZM2G140996 3.34 28.19 8.43 o-methyltransferase zrp4
32 GRMZM2G046618 0.86 6.99 8.16 pectin methylesterase GRMZM2G316789 6.5 51.17 7.87 short-chain type GRMZM2G006937 108.21 834.78 7.71 cycloartenol synthase GRMZM5G803503 24.87 184.98 7.44 ribosomal protein s19 GRMZM2G356076 1.23 8.75 7.13 brassinosteroid insensitive 1 precursor GRMZM2G150374 8.47 60.12 7.1 nodulin 21 -like transporter family protein GRMZM2G043336 6.52 46.19 7.09 jasmonate induced protein GRMZM2G077436 1.62 11.23 6.93 rna polymerase sigma factor GRMZM2G326933 7.07 48.07 6.8 hypothetical protein [Zea mays] GRMZM2G427404 12.48 84.51 6.77 ribosomal protein l2 GRMZM2G011896 1.82 10.87 5.96 leucine-rich receptor-like protein kinase GRMZM2G454056 3.04 17.87 5.88 hypothetical protein LOC100274968 [Zea mays] GRMZM2G087243 1.61 9.14 5.68 armadillo beta-catenin-like repeat family expressed GRMZM2G013430 7.04 39.69 5.64 serine acetyltransferase mitochondrial GRMZM2G175860 14.47 77.87 5.38 dnaj protein GRMZM2G151227 1.19 6.34 5.34 chalcone synthase GRMZM2G169013 20.88 109.78 5.26 hypothetical protein LOC100382353 [Zea mays] GRMZM2G164318 4.11 21.14 5.14 beta-carotene hydroxylase 1 GRMZM2G050997 1.73 8.87 5.13 cytokinin oxidase GRMZM2G026927 2.68 13.47 5.02 ovate family protein 4 GRMZM2G121753 17.28 83.54 4.84 sant myb protein GRMZM2G179024 4.43 20.83 4.7 pseudo response regulator GRMZM2G156127 1.88 8.79 4.68 cytokinin-n-glucosyltransferase 1 GRMZM2G372074 144.09 674.02 4.68 kda proline-rich protein AC204212.4_FG001 19.61 91.14 4.65 rad-like 6 protein GRMZM2G137046 15.86 73.24 4.62 transcription factor hy5 GRMZM2G165972 2.89 13.03 4.51 hsf-type dna-binding domain containing expressed GRMZM2G125850 5.54 24.2 4.37 hypothetical protein LOC100279907 [Zea mays] GRMZM2G568636 17.9 75.97 4.25 nitrate reductase
33 GRMZM2G026833 8.02 34.02 4.24 golden2-like transcription factor AC189750.4_FG004 22.68 93.12 4.11 prh26 protein GRMZM2G034471 3.82 15.67 4.1 cytochrome p450 GRMZM2G171390 7.98 31.93 4 chlorophyllide a oxygenase GRMZM2G129451 8.61 34.3 3.98 soluble starch synthase i GRMZM2G087896 34.2 132.79 3.88 phosphatidic acid phosphatase-related protein GRMZM2G059634 7.09 27.02 3.81 duf26 domain-containing protein 1 GRMZM2G106921 2.21 8.08 3.66 tellurite resistance protein GRMZM2G124963 18.98 69.02 3.64 alanine aminotransferase GRMZM2G157679 3.29 11.33 3.44 gras family transcription factor GRMZM5G859388 10.81 36.94 3.42 rpl32 GRMZM2G041774 10.73 35.83 3.34 hypothetical protein SORBIDRAFT_03g032140 [Sorghum bicolor GRMZM2G136081 13.07 42.97 3.29 plant-specific domain tigr01615 family protein GRMZM2G102959 7.22 23.72 3.28 nitrite reductase GRMZM2G357636 30.08 98.52 3.27 syntaxin 132 GRMZM2G172834 40.52 130.35 3.22 annexin a4 GRMZM5G895554 8.22 26.41 3.21 adenylyl-sulfate kinase 1 GRMZM2G047262 8.8 28.08 3.19 nucleoside nucleotide kinase domain-containing protein
34 TableS5 441 over-represented genes of 830 significantly up-regulated genes in heterozygous Rg1 leaves
FPKM FPKM Fold change GeneID P value value of Annotation value of WT (MT/WT) MT neurogenic locus notch GRMZM2G154036 0.00093 28.84 3.95 7.30 protein precursor-like ef hand family GRMZM2G312661 0.00093 40.07 8.64 4.64 expressed GRMZM2G358059 0.00093 57.19 18.32 3.12 calreticulin precursor GRMZM2G430600 0.00093 155.62 47.34 3.29 calmodulin GRMZM2G044963 0.00093 24.49 7.01 3.49 calmodulin GRMZM2G067511 0.00093 31.48 8.47 3.72 calmodulin xyloglucan endotransglycosylase GRMZM2G119783 6.10E-09 315.36 42.57 7.41 hydrolase protein 8 precursor xyloglucan GRMZM2G026980 6.10E-09 86.96 13.65 6.37 endotransglycosylase xyloglucan endotransglucosylase GRMZM2G413044 6.10E-09 42.12 0.98 43.20 hydrolase protein 32 precursor xyloglucan endotransglucosylase GRMZM2G113761 6.10E-09 22.46 4.03 5.57 hydrolase protein 32 precursor GRMZM2G165357 6.10E-09 123.88 26.03 4.76 udp-xyl synthase 6 udp-glucuronic acid GRMZM2G381473 6.10E-09 27.33 6.27 4.36 decarboxylase udp-glucuronic acid GRMZM2G007195 6.10E-09 32.05 7.85 4.08 decarboxylase GRMZM2G007404 6.10E-09 31.99 8.74 3.66 udp-glucuronic acid
35 decarboxylase trehalose-6-phosphate GRMZM2G312521 6.10E-09 170.38 43.25 3.94 synthase trehalose-6-phosphate *GRMZM2G416836 6.10E-09 17.31 synthase trehalose 6-phosphate GRMZM2G527891 6.10E-09 9.91 2.71 3.65 synthase trehalose 6-phosphate GRMZM2G008226 6.10E-09 22.37 5.66 3.95 synthase trehalose 6-phosphate GRMZM2G366659 6.10E-09 20.29 3.77 5.38 synthase GRMZM2G152908 6.10E-09 112.90 35.92 3.14 sucrose synthase GRMZM2G062156 6.10E-09 52.50 16.90 3.11 sorbitol transporter reversibly glycosylated GRMZM2G073725 6.10E-09 139.12 33.51 4.15 polypeptide reversibly glycosylated GRMZM2G087326 6.10E-09 38.76 4.74 8.18 polypeptide GRMZM2G388585 6.10E-09 18.31 4.76 3.84 related receptor serine threonine AC193632.2_FG002 6.10E-09 9.80 0.86 11.43 kinase pr5k-like GRMZM2G066290 6.10E-09 13.10 3.71 3.53 pyruvate kinase pyrophosphate- dependent GRMZM2G450163 6.10E-09 50.22 7.59 6.62 phosphofructokinase alpha subunit polygalacturonase-like GRMZM2G107073 6.10E-09 23.61 1.75 13.46 protein() GRMZM5G863596 6.10E-09 34.88 8.18 4.26 plastid alpha-amylase non-cell-autonomous GRMZM2G041356 6.10E-09 36.02 2.46 14.65 protein pathway2 plasmodesmal receptor GRMZM2G126900 6.10E-09 275.47 22.09 12.47 myo-inositol oxygenase GRMZM2G178025 6.10E-09 11.49 3.76 3.05 endo-1,4-beta-glucanase AC199765.4_FG008 6.10E-09 20.82 6.47 3.22 endo-1,4-beta-glucanase
36 mannan endo- -beta- *GRMZM2G471065 6.10E-09 9.82 mannosidase 6 GRMZM2G102183 6.10E-09 24.04 0.39 62.18 malate synthase GRMZM2G137535 6.10E-09 620.52 31.42 19.75 lichenase-2 precursor GRMZM2G128929 6.10E-09 13.44 1.24 10.84 lactate dehydrogenase ketose-bisphosphate GRMZM2G431708 6.10E-09 15.04 3.93 3.83 aldolase class-ii-like protein glycosyl hydrolase family 3 c terminal GRMZM2G035503 6.10E-09 18.23 3.88 4.69 domain containing expressed glycosyl hydrolase GRMZM2G002260 6.10E-09 8.30 0.31 27.17 family 10 protein glucan endo-1,3-beta- GRMZM2G019619 6.10E-09 81.30 22.78 3.57 glucosidase 7 precursor glucan endo-1,3-beta- GRMZM5G805609 6.10E-09 50.43 3.43 14.72 glucosidase 7 precursor GRMZM2G051677 6.10E-09 172.40 45.45 3.79 fructokinase-2 exocyst complex subunit GRMZM2G144739 6.10E-09 5.49 0.72 7.59 sec15-like family protein putative endo-beta-1,4- GRMZM2G451965 6.10E-09 26.85 6.36 4.22 glucanase precursor GRMZM2G003794 6.10E-09 12.11 2.65 4.57 endo- -beta-xylanase GRMZM2G154678 6.10E-09 11.36 2.54 4.47 endo-1,4-beta-glucanase dolichyl- diphosphooligosaccharid GRMZM2G462325 6.10E-09 28.77 8.41 3.42 e--protein glycosyltransferase 48 kdasubunit precursor GRMZM2G129189 6.10E-09 146.81 27.65 5.31 class iv chitinase class iii chitinase GRMZM2G162359 6.10E-09 40.22 4.29 9.39 homologue ( hib3h-a)
37 *GRMZM2G358153 6.10E-09 19.93 chitinase 1 *GRMZM2G005633 6.10E-09 14.70 chitinase *GRMZM2G051921 6.10E-09 3.30 chitinase GRMZM2G051943 6.10E-09 474.57 31.35 15.14 chitinase GRMZM2G145518 6.10E-09 30.89 1.42 21.80 chitinase GRMZM2G122431 6.10E-09 7.93 0.91 8.75 cellulose synthase isolog GRMZM2G424832 6.10E-09 68.97 22.57 3.06 cellulose synthase cellulase containing *GRMZM2G147221 6.10E-09 14.69 protein calcium-dependent GRMZM2G099425 6.10E-09 5.61 0.92 6.08 protein GRMZM5G810727 6.10E-09 51.37 8.32 6.18 beta-glucosidase beta-galactosidase GRMZM2G127123 6.10E-09 50.21 13.71 3.66 expressed GRMZM2G038281 6.10E-09 18.17 4.44 4.09 beta-galactosidase 10 GRMZM2G058310 6.10E-09 21.13 4.78 4.42 beta-amylase beta-1,3-glucanase GRMZM2G125032 6.10E-09 277.37 9.54 29.08 precursor putative beta-1,3- GRMZM2G431039 6.10E-09 52.03 6.27 8.30 glucanase GRMZM2G065585 6.10E-09 14.97 0.90 16.57 beta-1,3 glucanase GRMZM2G095126 6.10E-09 185.65 13.89 13.36 alpha-galactosidase *GRMZM2G138468 6.10E-09 19.35 alpha-amylase 6-phosphogluconate GRMZM2G145715 6.10E-09 53.48 14.59 3.67 dehydrogenase 6-phosphogluconate GRMZM2G127798 6.10E-09 96.33 30.15 3.20 dehydrogenase 2-oxoglutarate e1 GRMZM2G151169 6.10E-09 6.81 1.17 5.80 component *GRMZM2G465046 2.10E-06 10.01 zinc finger xylem serine proteinase GRMZM2G115773 2.10E-06 11.17 2.76 4.05 1 GRMZM2G054332 2.10E-06 13.01 0.33 39.23 abc transporter family
38 GRMZM2G157564 2.10E-06 79.18 18.14 4.36 abc transporter family GRMZM5G817964 2.10E-06 23.54 2.42 9.72 abc transporter family *GRMZM2G082118 2.10E-06 4.46 vq motif family protein vacuolar proton- GRMZM2G148200 2.10E-06 19.74 4.38 4.51 inorganic pyrophosphatase u-box domain- GRMZM2G115000 2.10E-06 20.12 5.95 3.38 containing protein 7 ubiquitin-conjugating GRMZM2G010460 2.10E-06 55.46 17.06 3.25 enzyme -like ubiquitin-conjugating *GRMZM2G000601 2.10E-06 5.74 enzyme e2-17 kda ubiquitin carboxyl- GRMZM2G113821 2.10E-06 131.99 16.43 8.03 terminal hydrolase 4 tyrosine dopa *GRMZM2G093125 2.10E-06 8.35 decarboxylase GRMZM2G124759 2.10E-06 1263.16 141.56 8.92 triacylglycerol lipase tartrate-resistant acid *GRMZM2G141584 2.10E-06 11.83 phosphatase type 5 precursor superkiller viralicidic GRMZM2G047949 2.10E-06 24.74 6.24 3.96 activity 2-like 2-like GRMZM2G143494 2.10E-06 74.51 20.11 3.70 strictosidine synthase soluble inorganic GRMZM2G026470 2.10E-06 137.50 24.77 5.55 pyrophosphatase skip interacting protein *GRMZM2G703021 2.10E-06 27.50 3 serine carboxypeptidase GRMZM2G452896 2.10E-06 16.46 5.45 3.02 s28 family protein serine carboxypeptidase GRMZM2G020146 2.10E-06 9.62 2.49 3.86 iii precursor *GRMZM2G126541 2.10E-06 8.01 serine carboxypeptidase *GRMZM2G041732 2.10E-06 19.74 rna helicase *GRMZM2G376085 2.10E-06 6.57 ring u-box domain and
39 arm repeat-containing protein purple acid phosphatase GRMZM2G134054 2.10E-06 11.77 1.56 7.53 isoform b purple acid phosphatase GRMZM2G109405 2.10E-06 20.13 3.53 5.71 29 GRMZM2G073860 2.10E-06 51.40 14.24 3.61 purple acid phosphatase *GRMZM2G434170 2.10E-06 8.72 purple acid phosphatase protein phosphatase 2c GRMZM2G071087 2.10E-06 217.00 68.81 3.15 containing expressed GRMZM2G107565 2.10E-06 9.90 2.79 3.54 protein phosphatase GRMZM2G040452 2.10E-06 162.50 37.40 4.34 protein phosphatase GRMZM2G130943 2.10E-06 12.15 2.17 5.60 protein phosphatase GRMZM2G144109 2.10E-06 46.31 10.37 4.47 protein phosphatase proteasome subunit GRMZM2G050467 2.10E-06 47.08 13.42 3.51 alpha type 7 GRMZM2G153208 2.10E-06 22.75 3.20 7.12 pr17c precursor GRMZM2G054559 2.10E-06 34.13 8.43 4.05 phospholipase d GRMZM2G137435 2.10E-06 37.57 4.76 7.89 phospholipase c phenylalanine ammonia- GRMZM2G081582 2.10E-06 44.54 13.00 3.43 lyase GRMZM2G128682 2.10E-06 7.80 0.97 8.02 pectinesterase 41 GRMZM2G167637 2.10E-06 25.13 5.84 4.30 pectin methylesterase *GRMZM2G138355 2.10E-06 15.46 nudix hydrolase 13 nodulin 21 family GRMZM2G108537 2.10E-06 37.54 9.29 4.04 expressed GRMZM2G138624 2.10E-06 13.58 2.46 5.53 nicotinamidase 1 GRMZM2G128880 2.10E-06 16.11 3.40 4.74 nicotinamidase 1 neutral alkaline GRMZM2G118737 2.10E-06 17.44 4.26 4.09 invertase multiple inositol GRMZM2G151967 2.10E-06 27.32 5.38 5.08 polyphosphate phosphatase a1
40 GRMZM2G004748 2.10E-06 17.76 4.22 4.21 abc transporter family abc transporter C family GRMZM2G361256 2.10E-06 19.75 3.69 5.35 member 3 metal ion binding *GRMZM2G028134 2.10E-06 7.02 protein GRMZM2G055698 2.10E-06 71.92 21.93 3.28 membrane protein GRMZM2G109560 2.10E-06 62.60 16.66 3.76 lon protease GRMZM2G059693 2.10E-06 19.38 3.42 5.66 lipase class 3-like leucine-rich repeat- GRMZM2G173647 2.10E-06 13.39 3.92 3.42 containing protein leucine rich repeat GRMZM2G044724 2.10E-06 43.38 12.47 3.48 family expressed leucine rich repeat GRMZM2G443525 2.10E-06 65.95 16.99 3.88 family expressed *AC214360.3_FG001 2.10E-06 30.08 iso-kaurene synthase hypothetical protein GRMZM2G132331 2.10E-06 25.66 2.74 9.36 SORBIDRAFT_10g024 300 [Sorghum bicolor] GRMZM2G363813 2.10E-06 8.86 2.34 3.78 hipl1 protein heavy metal transporter GRMZM2G175576 2.10E-06 15.62 2.12 7.37 3 haloacid dehalogenase- GRMZM2G179432 2.10E-06 760.94 227.28 3.35 like hydrolase domain- containing protein GRMZM2G023436 2.10E-06 54.04 9.40 5.75 gtp-binding protein GRMZM2G024104 2.10E-06 43.90 2.24 19.64 glutamine synthetase GRMZM5G826838 2.10E-06 26.44 4.40 6.01 glutamate decarboxylase *GRMZM2G067919 2.10E-06 4.36 globulin 1 gibberellin receptor GRMZM2G156310 2.10E-06 28.02 6.19 4.53 gid1l2 gdp-mannose GRMZM2G074850 2.10E-06 22.68 4.68 4.84 dehydratase 2 GRMZM2G010823 2.10E-06 25.06 6.25 4.01 fumarate hydratase 2 GRMZM2G118917 2.10E-06 18.07 5.64 3.20 exosome complex
41 exonuclease rrp6 GRMZM2G047139 2.10E-06 54.85 3.71 14.78 esterase precursor GRMZM2G148460 2.10E-06 17.29 5.04 3.43 esterase precursor GRMZM2G085176 2.10E-06 43.66 4.33 10.09 esterase precursor GRMZM2G172230 2.10E-06 50.54 7.86 6.43 erd1 protein eps15 homology domain GRMZM2G113613 2.10E-06 86.82 16.51 5.26 1 protein endoplasmic reticulum- GRMZM2G141704 2.10E-06 10.62 1.94 5.47 type calcium- transporting atpase 4 dynein light chain GRMZM2G016435 2.10E-06 41.18 5.15 7.99 flagellar outer arm dynein light chain GRMZM2G346133 2.10E-06 124.73 33.77 3.69 flagellar outer arm dynamin-related protein GRMZM2G055883 2.10E-06 39.05 5.15 7.58 expressed dynamin-related protein GRMZM2G145827 2.10E-06 27.45 8.45 3.25 1c dna-3-methyladenine GRMZM2G132303 2.10E-06 26.87 3.04 8.83 glycosylase i *GRMZM2G160473 2.10E-06 10.55 dicer-like protein 4 AC191097.3_FG007 2.10E-06 52.94 12.12 4.37 cytochrome b561 GRMZM2G456217 2.10E-06 48.10 9.14 5.26 cysteine proteinase GRMZM2G006937 2.10E-06 33.07 10.32 3.21 cycloartenol synthase GRMZM5G855337 2.10E-06 17.29 1.90 9.10 ctp synthase GRMZM2G132547 2.10E-06 78.97 10.20 7.74 ctp synthase GRMZM2G330302 2.10E-06 28.64 3.55 8.07 ctp synthase GRMZM2G124365 2.10E-06 12.41 1.75 7.11 chorismate mutase GRMZM2G113191 2.10E-06 11.37 1.89 6.03 carbonic anhydrase bifunctional nuclease GRMZM2G168744 2.10E-06 12.35 2.31 5.36 bfn1 GRMZM2G332809 2.10E-06 7.33 1.95 3.76 beta-tubulin GRMZM2G066191 2.10E-06 32.81 8.36 3.93 beta-4 tubulin
42 GRMZM2G133802 2.10E-06 93.33 7.92 11.79 beta chain GRMZM2G177812 2.10E-06 148.03 36.93 4.01 atp-binding cassette aspartyl GRMZM2G071871 2.10E-06 24.31 5.82 4.18 transcarbamylase aspartyl protease family GRMZM2G168115 2.10E-06 21.66 2.60 8.32 protein GRMZM2G078472 2.10E-06 15.74 0.54 28.91 asparagine synthetase GRMZM2G053669 2.10E-06 214.48 2.81 76.44 asparagine synthetase anter-specific proline- GRMZM2G101117 2.10E-06 60.01 17.01 3.53 rich protein apg ankyrin repeat-rich GRMZM2G315072 2.10E-06 35.06 7.48 4.69 protein GRMZM2G152417 2.10E-06 15.81 1.22 12.95 amp dependent GRMZM2G074759 2.10E-06 21.70 4.86 4.47 amp dependent alpha-l-fucosidase 2 GRMZM2G052562 2.10E-06 138.18 19.78 6.98 precursor GRMZM2G448523 2.10E-06 117.38 27.71 4.24 aig1 family expressed *GRMZM5G812228 2.10E-06 16.73 acyl- synthetase abc transporter family GRMZM2G099619 2.10E-06 25.59 0.90 28.46 protein abc transporter family GRMZM2G032218 2.10E-06 7.86 2.11 3.73 member 9 GRMZM2G308860 2.10E-06 32.32 8.86 3.65 abc transporter GRMZM2G444560 2.10E-06 21.90 0.64 34.15 aaa-type atpase-like wound-induced serine *GRMZM2G156632 5.50E-07 852.02 protease inhibitor phytosulfokines 2 GRMZM2G031317 5.50E-07 28.41 6.68 4.26 precursor peroxidase family GRMZM2G341934 5.50E-07 24.39 7.42 3.29 expressed *GRMZM2G061230 5.50E-07 18.26 peroxidase pathogenesis-related GRMZM2G465226 5.50E-07 36.97 7.91 4.67 protein class i *GRMZM2G336337 5.50E-07 4.45 laccase
43 GRMZM2G126975 5.50E-07 11.98 1.30 9.18 hypothetical protein GRMZM2G045809 5.50E-07 16.45 3.26 5.04 germin protein type 1 *GRMZM2G166141 5.50E-07 47.16 germin protein type 1 expansin-like 3 GRMZM2G026147 5.50E-07 208.66 66.41 3.14 precursor expansin-like 3 GRMZM2G127072 5.50E-07 91.82 20.10 4.57 precursor *GRMZM2G094523 5.50E-07 11.48 expansin os-expa3 GRMZM2G339122 5.50E-07 346.26 13.25 26.13 expansin os-expa2 GRMZM2G361064 5.50E-07 300.56 41.22 7.29 expansin os-expa2 bowman-birk type wound-induced GRMZM2G011523 5.50E-07 735.04 105.19 6.99 proteinase inhibitor wip1 precursor bowman-birk type GRMZM2G075315 5.50E-07 47.70 2.28 20.94 trypsin inhibitor bowman-birk type GRMZM2G055802 5.50E-07 35.39 3.40 10.42 trypsin inhibitor beta-expansin 4 GRMZM2G056236 5.50E-07 26.59 0.64 41.56 precursor beta-expansin 4 *GRMZM2G327266 5.50E-07 9.45 precursor GRMZM2G169967 5.50E-07 45.11 11.15 4.04 beta-expansin 3 beta-expansin 1a GRMZM2G094990 5.50E-07 424.00 55.13 7.69 precursor beta-expansin 1a GRMZM2G176595 5.50E-07 224.20 15.03 14.92 precursor beta-expansin 1a GRMZM2G013002 5.50E-07 165.63 5.74 28.85 expressed beta-expansin 1a GRMZM2G082520 5.50E-07 259.15 9.47 27.36 expressed beta-expansin 1a *GRMZM2G342246 5.50E-07 23.87 expressed GRMZM2G021621 5.50E-07 80.25 12.22 6.57 beta-expansin 1a
44 expressed beta-expansin 1a *GRMZM2G021427 5.50E-07 9.80 expressed alpha-expansin 6 GRMZM2G368886 5.50E-07 14.47 1.35 10.73 precursor heparanase-like protein GRMZM2G098397 1.20E-07 11.97 3.02 3.96 2 precursor transmembrane GRMZM2G028521 1.40E-06 258.60 63.92 4.05 expressed cinnamate-4- GRMZM2G147245 1.40E-06 12.21 3.21 3.80 hydroxylase rna-dependent rna GRMZM2G481730 1.40E-06 12.32 2.97 4.15 polymerase ph domain containing GRMZM2G122267 1.40E-06 77.93 6.22 12.54 protein GRMZM2G042347 1.40E-06 22.40 2.93 7.65 peroxidase precursor GRMZM2G126261 1.40E-06 45.39 7.42 6.12 peroxidase poc1 GRMZM2G176085 1.40E-06 71.49 18.84 3.79 peroxidase 73 AC197758.3_FG004 1.40E-06 49.91 2.63 18.99 peroxidase 52 *GRMZM2G095404 1.40E-06 20.15 peroxidase 49 precursor GRMZM2G085967 1.40E-06 78.62 4.28 18.35 peroxidase 3 GRMZM2G138450 1.40E-06 10.34 1.26 8.20 peroxidase 27 GRMZM2G080183 1.40E-06 8.71 0.91 9.58 peroxidase 16 GRMZM2G103342 1.40E-06 117.10 17.36 6.75 peroxidase 12 GRMZM2G321839 1.40E-06 35.69 6.44 5.55 peroxidase 1 precursor GRMZM2G122853 1.40E-06 21.53 4.02 5.36 peroxidase 1 GRMZM2G135108 1.40E-06 32.22 8.48 3.80 peroxidase GRMZM2G450233 1.40E-06 554.52 109.08 5.08 peroxidase GRMZM2G427815 1.40E-06 17.06 1.71 9.95 peroxidase GRMZM2G441541 1.40E-06 114.23 25.90 4.41 nadph oxidase medium chain fatty acid GRMZM2G132478 1.40E-06 7.04 1.13 6.23 hydroxylase GRMZM5G822593 1.40E-06 11.79 1.14 10.34 lipoxygenase
45 GRMZM2G109130 1.40E-06 75.22 5.68 13.25 lipoxygenase 1 GRMZM2G009479 1.40E-06 55.93 14.51 3.86 lipoxygenase GRMZM2G156861 1.40E-06 933.81 176.59 5.29 lipoxygenase GRMZM2G109056 1.40E-06 31.84 5.06 6.29 lipoxygenase iron ascorbate- *GRMZM2G162158 1.40E-06 7.25 dependent oxidoreductase GRMZM2G083526 1.40E-06 167.03 35.08 4.76 gl1 protein GRMZM2G029912 1.40E-06 168.36 18.21 9.24 gl1 protein GRMZM2G427618 1.40E-06 19.73 5.30 3.73 gibberellin 2-oxidase GRMZM2G025832 1.40E-06 10.26 0.72 14.23 flavonoid 3-hydroxylase AC210173.4_FG005 1.40E-06 10.10 1.83 5.52 ferulate-5-hydroxylase elicitor-inducible GRMZM2G431288 1.40E-06 7.31 0.71 10.33 cytochrome p450 GRMZM2G144668 1.40E-06 32.33 8.13 3.98 desacetoxyvindoline GRMZM2G349187 1.40E-06 14.96 1.51 9.88 cytochrome p450 GRMZM2G056247 1.40E-06 13.42 2.98 4.50 cytochrome p450 GRMZM2G020761 1.40E-06 40.13 10.12 3.96 cytochrome p450 GRMZM2G066441 1.40E-06 14.88 3.28 4.54 cytochrome p450 *GRMZM2G167673 1.40E-06 9.39 cytochrome p450 *GRMZM2G075255 1.40E-06 12.03 cer1 protein GRMZM2G141473 1.40E-06 35.71 10.71 3.33 aldehyde oxidase GRMZM2G123667 1.40E-06 12.11 2.62 4.62 5 protein 2og-fe oxygenase family *GRMZM2G363893 1.40E-06 14.56 expressed 1-aminocyclopropane-1- GRMZM2G007249 1.40E-06 15.01 3.43 4.38 carboxylate oxidase 1-aminocyclopropane-1- GRMZM2G126732 1.40E-06 18.83 5.20 3.62 carboxylate oxidase GRMZM2G118821 0.00022 58.34 11.47 5.09 zip transporter receptor-type protein AC194974.3_FG005 0.00022 6.11 1.15 5.32 kinase lrk1 GRMZM2G142063 0.00022 183.68 58.84 3.12 inositol transporter 2
46 histidine amino acid GRMZM2G149216 0.00022 17.58 2.92 6.02 transporter GRMZM2G332562 0.00022 6.53 0.68 9.59 amino acid transporter GRMZM2G180659 0.00022 153.88 21.96 7.01 amino acid GRMZM2G046743 0.00022 17.84 4.65 3.84 amino acid GRMZM2G036448 0.00022 205.71 43.83 4.69 amino acid GRMZM2G022642 2.40E-06 14.55 3.03 4.81 amino acid transporter GRMZM2G050984 7.50E-07 8.82 2.18 4.05 xanthine dehydrogenase udp-glucose 6- GRMZM5G862540 7.50E-07 104.39 20.73 5.03 expressed udp-glucose 6- GRMZM2G328500 7.50E-07 143.70 20.07 7.16 expressed GRMZM2G152258 7.50E-07 30.40 5.60 5.42 tropinone reductase 2 stearoyl-acyl carrier GRMZM2G119305 7.50E-07 39.56 1.04 38.10 protein desaturase short chain dehydrogenase GRMZM2G128577 7.50E-07 72.74 22.90 3.18 reductase family expressed GRMZM2G052266 7.50E-07 12.54 0.91 13.81 sarcosine oxidase rossmann-fold nad GRMZM2G135277 7.50E-07 23.39 5.98 3.91 -binding domain- containing protein GRMZM2G304362 7.50E-07 23.87 3.95 6.04 ribonucleotide reductase GRMZM2G053720 7.50E-07 129.84 22.76 5.71 proline oxidase GRMZM2G153799 7.50E-07 31.58 3.58 8.82 polyamine oxidase GRMZM2G071343 7.50E-07 43.59 7.77 5.61 polyamine oxidase nitrate reductase GRMZM2G154146 7.50E-07 15.57 2.18 7.14 apoenzyme nadp-specific isocitrate GRMZM2G033515 7.50E-07 37.91 5.77 6.57 dehydrogenase nad h-dependent GRMZM2G126062 7.50E-07 6.41 1.28 5.02 oxidoreductase GRMZM2G134708 7.50E-07 20.70 4.34 4.77 monodehydroascorbate
47 reductase mitochondrial aldehyde GRMZM2G125268 7.50E-07 40.94 5.20 7.87 dehydrogenase GRMZM2G379540 7.50E-07 86.27 27.66 3.12 membrane protein malate oxidoreductase GRMZM2G406672 7.50E-07 20.15 6.34 3.18 (malic enzyme) lysine-ketoglutarate reductase saccharopine GRMZM2G181362 7.50E-07 76.95 9.92 7.76 dehydrogenase bifunctional enzyme isovaleryl- GRMZM2G052630 7.50E-07 10.76 2.76 3.90 dehydrogenase GRMZM5G853854 7.50E-07 37.66 8.07 4.67 glycolate oxidase glutamate GRMZM2G427097 7.50E-07 25.91 2.59 10.00 dehydrogenase 2 gibberellin 2-beta- GRMZM2G006964 7.50E-07 11.77 1.43 8.21 dioxygenase 7 GRMZM2G025536 7.50E-07 9.70 1.77 5.47 fe2+ dioxygenase-like d-isomer specific 2- GRMZM2G159587 7.50E-07 7.49 0.85 8.77 hydroxyacid dehydrogenase GRMZM2G119116 7.50E-07 23.62 4.29 5.51 desacetoxyvindoline 4- GRMZM2G033555 7.50E-07 8.18 1.18 6.95 cinnamoyl- reductase GRMZM2G354909 7.50E-07 117.29 11.43 10.26 carbonyl reductase 3 aldehyde dimeric nadp- GRMZM2G118800 7.50E-07 44.11 7.43 5.94 preferring GRMZM2G090087 7.50E-07 57.00 16.79 3.40 aldehyde dehydrogenase GRMZM2G442658 7.50E-07 48.08 15.14 3.17 alcohol dehydrogenase 1 GRMZM2G154007 7.50E-07 40.18 2.24 17.92 alcohol dehydrogenase GRMZM2G152975 7.50E-07 30.52 7.25 4.21 alcohol dehydrogenase GRMZM2G098346 7.50E-07 121.03 20.08 6.03 alcohol dehydrogenase GRMZM5G854613 7.50E-07 65.65 17.50 3.75 aim1 protein GRMZM2G459755 7.50E-07 21.25 5.14 4.14 aim1 protein
48 GRMZM2G035741 7.50E-07 453.80 133.67 3.39 91a protein 2-oxoisovalerate GRMZM2G088689 7.50E-07 33.73 6.26 5.39 dehydrogenase e1 alpha subunit 2og-fe oxygenase family GRMZM2G475380 7.50E-07 78.28 4.93 15.87 expressed 1-deoxy-d-xylulose 5- *GRMZM2G036290 7.50E-07 15.01 phosphate reductoisomerase 1-aminocyclopropane-1- AC148152.3_FG005 7.50E-07 35.38 0.64 55.39 carboxylate oxidase vacuolar atp synthase 16 GRMZM2G047476 4.90E-05 198.23 51.56 3.84 kda proteolipid subunit *GRMZM2G042933 4.90E-05 11.23 amino acid permease transparent testa 12 GRMZM2G161809 7.00E-05 9.00 2.14 4.20 protein ripening regulated GRMZM2G166459 7.00E-05 9.70 1.92 5.06 protein harpin-induced protein 1 GRMZM2G094589 7.00E-05 87.35 19.47 4.49 containing expressed GRMZM2G079440 7.00E-05 166.80 5.34 31.26 dehydrin7 GRMZM2G373522 7.00E-05 238.98 23.99 9.96 dehydrin cor410 calcineurin b-like GRMZM2G116584 7.00E-05 56.61 12.34 4.59 protein 10 auxin response GRMZM2G028980 7.00E-05 35.60 5.96 5.98 transcription factor GRMZM2G317900 7.00E-05 9.20 1.35 6.83 auxin response factor 7a GRMZM2G035405 7.00E-05 33.27 9.17 3.63 auxin response factor xyloglucan GRMZM2G015983 2.10E-06 31.39 5.36 5.86 fucosyltransferase GRMZM2G145109 2.10E-06 6.66 0.77 8.65 wall-associated kinase 4 GRMZM2G077655 2.10E-06 9.60 1.40 6.85 wall-associated kinase 4 GRMZM2G014560 2.10E-06 11.16 1.44 7.74 wall-associated kinase 4 GRMZM2G145045 2.10E-06 10.33 0.75 13.79 wall-associated kinase 4
49 undecaprenyl GRMZM2G054803 2.10E-06 31.16 6.19 5.03 pyrophosphate synthetase udp-n- GRMZM2G044629 2.10E-06 68.81 19.42 3.54 acetylglucosamine pyrophosphorylase udp-n- GRMZM2G019986 2.10E-06 33.44 10.59 3.16 acetylglucosamine pyrophosphorylase udp-glycosyltransferase- GRMZM2G173315 2.10E-06 53.72 7.20 7.46 like protein udp-glycosyltransferase- GRMZM2G337048 2.10E-06 25.31 4.81 5.26 like protein udp-glycosyltransferase- *GRMZM2G021786 2.10E-06 19.48 like protein transferring glycosyl GRMZM2G026889 2.10E-06 10.33 1.50 6.87 groups transferase family GRMZM2G061806 2.10E-06 10.14 0.75 13.50 expressed GRMZM2G313649 2.10E-06 5.66 0.75 7.53 tp53 regulating kinase t7 bacteriophage-type GRMZM2G381395 2.10E-06 36.77 7.23 5.09 single subunit rna polymerase strubbelig-receptor GRMZM2G162531 2.10E-06 46.26 9.44 4.90 family 8 protein steroid sulfotransferase GRMZM2G152179 2.10E-06 14.85 4.04 3.68 3 GRMZM2G334592 2.10E-06 14.98 2.08 7.20 s-receptor kinase somatic embryogenesis GRMZM2G384439 2.10E-06 92.81 19.76 4.70 receptor-like kinase 1 s-locus lectin protein GRMZM2G411668 2.10E-06 71.54 18.15 3.94 kinase family protein serine-threonine protein GRMZM2G007477 2.10E-06 26.83 0.73 36.62 plant- GRMZM2G079583 2.10E-06 28.29 0.68 41.55 serine threonine-specific
50 protein kinase npk15- like serine threonine-specific GRMZM2G059497 2.10E-06 25.63 4.68 5.47 protein kinase npk15- like serine threonine-specific *GRMZM2G431018 2.10E-06 7.70 protein kinase serine threonine-protein GRMZM2G029242 2.10E-06 28.50 7.23 3.94 kinase receptor precursor serine threonine-protein GRMZM2G101687 2.10E-06 8.52 1.79 4.75 kinase nak serine threonine-protein GRMZM2G081915 2.10E-06 40.40 11.95 3.38 kinase serine threonine-protein GRMZM2G316474 2.10E-06 29.15 7.73 3.77 kinase serine threonine protein GRMZM2G094541 2.10E-06 17.57 3.31 5.31 kinase serine threonine kinase GRMZM2G075438 2.10E-06 21.49 1.82 11.83 -related protein serine threonine kinase GRMZM2G076943 2.10E-06 9.43 1.02 9.21 protein serine threonine kinase *GRMZM5G856011 2.10E-06 10.46 protein rust resistance kinase GRMZM2G079219 2.10E-06 24.17 2.58 9.35 Lr10 rust resistance kinase GRMZM2G034611 2.10E-06 30.81 2.61 11.82 Lr10 rust resistance kinase GRMZM2G077914 2.10E-06 13.38 1.06 12.58 Lr10 rust resistance kinase GRMZM2G113421 2.10E-06 23.38 0.74 31.79 Lr10 rna polymerase iv GRMZM2G054225 2.10E-06 15.27 2.32 6.58 second largest subunit GRMZM2G474153 2.10E-06 9.96 1.58 6.32 receptor-type protein
51 kinase lrk1 receptor-like protein *GRMZM2G309380 2.10E-06 29.21 kinase 4 receptor-like protein GRMZM2G328785 2.10E-06 7.03 0.64 10.96 kinase GRMZM2G000620 2.10E-06 46.28 5.60 8.26 receptor-like kinase GRMZM2G317770 2.10E-06 31.85 10.18 3.13 receptor-like kinase receptor protein kinase- GRMZM2G333875 2.10E-06 23.76 6.22 3.82 like receptor protein kinase GRMZM2G358830 2.10E-06 8.63 1.78 4.85 perk1 receptor protein kinase GRMZM2G391794 2.10E-06 36.73 9.94 3.69 clavata1 GRMZM2G429714 2.10E-06 60.66 12.70 4.78 receptor protein protein kinase receptor GRMZM2G109830 2.10E-06 11.54 1.86 6.21 type precursor GRMZM2G026065 2.10E-06 10.05 2.47 4.07 protein kinase g11a protein kinase family GRMZM2G105415 2.10E-06 47.51 15.58 3.05 protein protein kinase domain- GRMZM2G113512 2.10E-06 20.84 5.59 3.73 containing protein protein kinase domain- GRMZM2G432796 2.10E-06 12.77 3.29 3.88 containing protein GRMZM2G177726 2.10E-06 2327.66 7.49 310.82 protein kinase GRMZM2G142544 2.10E-06 46.88 13.49 3.48 protein kinase GRMZM2G121565 2.10E-06 26.42 7.92 3.34 protein kinase GRMZM2G063069 2.10E-06 32.72 4.72 6.94 protein kinase GRMZM2G059740 2.10E-06 12.18 1.99 6.13 protein kinase *AC212835.3_FG008 2.10E-06 6.83 protein kinase GRMZM2G452142 2.10E-06 15.37 3.65 4.21 phytosulfokine receptor GRMZM2G104538 2.10E-06 40.94 10.91 3.75 phytol kinase phosphoenolpyruvate GRMZM2G135091 2.10E-06 11.34 2.54 4.47 carboxylase kinase
52 phosphatidylinositol 4- GRMZM2G114162 2.10E-06 29.14 9.49 3.07 kinase alpha phosphatidylinositol 3- GRMZM2G137558 2.10E-06 6.33 1.70 3.71 and 4-kinase family-like *GRMZM2G127418 2.10E-06 33.30 o-methyltransferase zrp4 *GRMZM2G099297 2.10E-06 5.28 o-methyltransferase zrp4 GRMZM2G017557 2.10E-06 18.87 5.06 3.73 o-methyltransferase zrp4 o-methyltransferase GRMZM2G085924 2.10E-06 11.53 1.11 10.37 family expressed GRMZM2G400470 2.10E-06 8.32 1.59 5.23 map kinase kinase GRMZM2G074262 2.10E-06 30.13 5.09 5.92 lstk-1-like kinase leucine-rich repeat GRMZM2G082191 2.10E-06 16.77 4.66 3.60 receptor-like protein kinase GRMZM2G466298 2.10E-06 29.38 2.06 14.25 lectin-like protein kinase *GRMZM2G142390 2.10E-06 6.85 kinase-like protein GRMZM5G802115 2.10E-06 10.79 2.80 3.85 kinase family protein GRMZM2G305822 2.10E-06 19.35 2.42 7.98 kinase family protein indole-3-acetate beta- *GRMZM5G896260 2.10E-06 31.81 glucosyltransferase hypothetical protein GRMZM2G410916 2.10E-06 39.78 9.16 4.34 SORBIDRAFT_03g008 020 hydroxycinnamoyl shikimate quinate *AC155610.2_FG007 2.10E-06 30.54 hydroxycinnamoyltransf erase-like protein hydroxycinnamoyl shikimate quinate GRMZM2G034360 2.10E-06 37.96 1.30 29.30 hydroxycinnamoyltransf erase-like protein hydroquinone GRMZM2G156026 2.10E-06 11.72 2.50 4.69 glucosyltransferase GRMZM2G098239 2.10E-06 256.10 58.17 4.40 hxxxd-type acyl-
53 transferase-like protein hxxxd-type acyl- GRMZM2G128564 2.10E-06 17.38 4.20 4.14 transferase-like protein histone-lysine n- GRMZM2G147619 2.10E-06 7.56 1.70 4.45 methyltransferase ashh1 histone-lysine n- h3 GRMZM2G025924 2.10E-06 5.32 0.49 10.84 lysine-9 specific suvh9 glycosyltransferase GRMZM2G105221 2.10E-06 33.15 1.71 19.35 family 61 protein glycosyltransferase GRMZM2G136580 2.10E-06 48.75 12.82 3.80 family 61 protein GRMZM2G076307 2.10E-06 14.23 1.59 8.97 glycosyl transferase glycerol-3-phosphate GRMZM2G083195 2.10E-06 67.22 6.82 9.86 acyltransferase 6 glycerol-3-phosphate GRMZM2G059637 2.10E-06 7.88 1.48 5.31 acyltransferase 5 GRMZM2G028821 2.10E-06 45.90 3.61 12.73 glutathione-s-transferase GRMZM2G064255 2.10E-06 267.74 78.43 3.41 glutathione-s-transferase *GRMZM2G025190 2.10E-06 10.11 glutathione-s-transferase GRMZM2G098890 2.10E-06 7.94 0.97 8.17 glucosyl transferase gdp-mannose GRMZM2G160506 2.10E-06 16.92 2.94 5.76 pyrophosphorylase flagellin-sensing 2-like GRMZM2G080041 2.10E-06 24.28 6.21 3.91 protein GRMZM2G445602 2.10E-06 593.61 117.19 5.07 fiddlehead-like protein fatty acid elongase 3- GRMZM2G104626 2.10E-06 61.28 18.34 3.34 ketoacyl- synthase 1 *GRMZM2G031790 2.10E-06 8.36 fatty acid elongase GRMZM2G089010 2.10E-06 22.53 3.51 6.43 ethylene receptor er glycerol-phosphate GRMZM2G166176 2.10E-06 15.82 3.96 4.00 acyltransferase dehydrodolichyl *GRMZM2G093146 2.10E-06 10.14 diphosphate synthase 6 *GRMZM2G082249 2.10E-06 8.98 cytokinin-o-
54 glucosyltransferase 3 GRMZM2G176206 2.10E-06 25.01 7.66 3.26 clavata1 receptor kinase cbl-interacting serine GRMZM2G051103 2.10E-06 27.95 8.03 3.48 threonine-protein kinase 9 cbl-interacting protein GRMZM2G013236 2.10E-06 88.18 19.76 4.46 kinase 23 cbl-interacting protein GRMZM2G113967 2.10E-06 111.28 22.18 5.02 kinase calcium-dependent GRMZM2G028926 2.10E-06 10.95 1.43 7.63 protein kinase calcium-dependent GRMZM2G320506 2.10E-06 21.05 6.37 3.31 protein kinase calcium-dependent GRMZM2G314396 2.10E-06 12.80 4.07 3.14 protein kinase Caffeoyl-CoA O- GRMZM2G127948 2.10E-06 219.11 59.22 3.70 methyltransferase 1 brassinosteroid insensitive 1-associated GRMZM2G125308 2.10E-06 24.38 7.50 3.25 receptor kinase 1 precursor brassinosteroid insensitive 1-associated GRMZM2G104760 2.10E-06 12.01 1.32 9.08 receptor kinase 1 precursor brassinosteroid insensitive 1-associated GRMZM5G867798 2.10E-06 17.87 5.72 3.12 receptor kinase 1 expressed branched-chain-amino- GRMZM2G153536 2.10E-06 23.32 0.97 23.98 acid aminotransferase chloroplast expressed GRMZM2G127141 2.10E-06 28.25 6.26 4.52 big map kinase benzoate carboxyl GRMZM2G126772 2.10E-06 84.24 2.13 39.59 methyltransferase
55 GRMZM5G852177 2.10E-06 16.61 2.39 6.94 atp binding GRMZM2G412885 2.10E-06 12.17 1.31 9.28 atp binding GRMZM2G052650 2.10E-06 17.62 3.55 4.96 atp binding GRMZM2G164974 2.10E-06 170.54 34.50 4.94 at1g68530 t26j14_10 AC233893.1_FG003 2.10E-06 84.36 18.60 4.54 at1g68530 t26j14_10 aspartate GRMZM2G094712 2.10E-06 20.02 4.71 4.25 aminotransferase anthranilate n- GRMZM2G035584 2.10E-06 149.51 45.19 3.31 benzoyltransferase protein 1 anthocyanidin 3-o- GRMZM2G061321 2.10E-06 13.68 3.76 3.64 glucosyltransferase GRMZM2G352855 2.10E-06 101.64 11.21 9.07 acyltransferase acetolactate synthase GRMZM2G407044 2.10E-06 27.61 1.54 17.96 amino acid binding protein 3-n-debenzoyl-2- GRMZM2G314898 2.10E-06 80.13 23.01 3.48 deoxytaxol n- benzoyltransferase 2-oxoisovalerate dehydrogenase e2 GRMZM2G033644 2.10E-06 44.71 14.82 3.02 component (dihydrolipoyl transacylase) 1-aminocyclopropane-1- GRMZM2G018006 2.10E-06 49.95 9.23 5.41 carboxylate synthase 10-deacetylbaccatin iii GRMZM2G129266 2.10E-06 12.39 1.70 7.27 10-o-acetyltransferase * indicates the genes are exclusively expressed in Rg1 leaves.
56 TableS6 Functional categories of 441 over-represented genes
Percentage of genes in each category Functional Categories accounting for 441genes (%) catalytic activity 16.72% transferase activity 6.40% oxidoreductase activity 4.15% transferase activity, transferring phosphorus-containing groups 3.80% phosphotransferase activity, alcohol group as acceptor 3.54% oxidation reduction 3.54% kinase activity 3.41% protein modification process 3.41% macromolecule modification 3.41% post-translational protein modification 3.37% phosphate metabolic process 3.37% phosphorus metabolic process 3.37% phosphorylation 3.28%
57 protein kinase activity 3.20% protein amino acid phosphorylation 3.15% protein serine/threonine kinase activity 3.07% carbohydrate metabolic process 2.81% iron ion binding 2.03% hydrolase activity, acting on glycosyl bonds 1.56% hydrolase activity, hydrolyzing O-glycosyl compounds 1.51% extracellular region 1.38% cellular carbohydrate metabolic process 1.34% response to chemical stimulus 1.12% polysaccharide metabolic process 0.86% calcium ion binding 0.86% transferase activity, transferring acyl groups 0.82% cell wall organization or biogenesis 0.78% transferase activity, transferring acyl groups other than amino-acyl groups 0.78% peroxidase activity 0.73% oxidoreductase activity, acting on peroxide as acceptor 0.73% oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor 0.73% antioxidant activity 0.73% oxidoreductase activity, acting on CH-OH group of donors 0.73% response to oxidative stress 0.69% reproduction 0.61% carbohydrate binding 0.61% cellular carbohydrate biosynthetic process 0.61% carbohydrate biosynthetic process 0.61% cellular polysaccharide metabolic process 0.56% organic acid transport 0.52% carboxylic acid transport 0.52%
58 sexual reproduction 0.48% polysaccharide catabolic process 0.30% plant-type cell wall organization or biogenesis 0.30% plant-type cell wall organization 0.30% chitinase activity 0.26% chitin metabolic process 0.26% chitin catabolic process 0.26% aminoglycan catabolic process 0.26% oxidoreductase activity, acting on single donors with incorporation of molecular oxygen 0.26% aminoglycan metabolic process 0.26% amino acid transport 0.26% amine transport 0.26% chitin binding 0.22% polysaccharide binding 0.22% pattern binding 0.22% lipoxygenase activity 0.22% oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, 0.22% incorporation of two atoms of oxygen
59