Eaves et al.
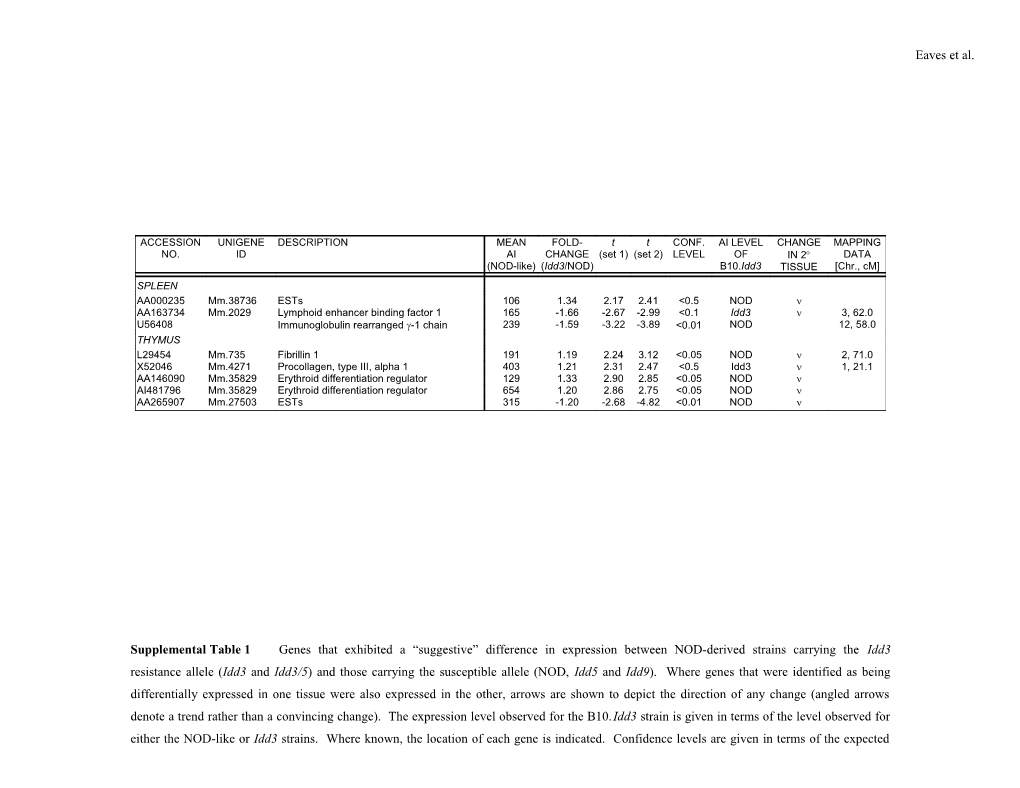
ACCESSION UNIGENE DESCRIPTION MEAN FOLD- t t CONF. AI LEVEL CHANGE MAPPING NO. ID AI CHANGE (set 1) (set 2) LEVEL OF IN 2 DATA (NOD-like) (Idd3/NOD) B10.Idd3 TISSUE [Chr., cM] SPLEEN AA000235 Mm.38736 ESTs 106 1.34 2.17 2.41 <0.5 NOD AA163734 Mm.2029 Lymphoid enhancer binding factor 1 165 -1.66 -2.67 -2.99 <0.1 Idd3 3, 62.0 U56408 Immunoglobulin rearranged -1 chain 239 -1.59 -3.22 -3.89 <0.01 NOD 12, 58.0 THYMUS L29454 Mm.735 Fibrillin 1 191 1.19 2.24 3.12 <0.05 NOD 2, 71.0 X52046 Mm.4271 Procollagen, type III, alpha 1 403 1.21 2.31 2.47 <0.5 Idd3 1, 21.1 AA146090 Mm.35829 Erythroid differentiation regulator 129 1.33 2.90 2.85 <0.05 NOD AI481796 Mm.35829 Erythroid differentiation regulator 654 1.20 2.86 2.75 <0.05 NOD AA265907 Mm.27503 ESTs 315 -1.20 -2.68 -4.82 <0.01 NOD
Supplemental Table 1 Genes that exhibited a “suggestive” difference in expression between NOD-derived strains carrying the Idd3 resistance allele (Idd3 and Idd3/5) and those carrying the susceptible allele (NOD, Idd5 and Idd9). Where genes that were identified as being differentially expressed in one tissue were also expressed in the other, arrows are shown to depict the direction of any change (angled arrows denote a trend rather than a convincing change). The expression level observed for the B10.Idd3 strain is given in terms of the level observed for either the NOD-like or Idd3 strains. Where known, the location of each gene is indicated. Confidence levels are given in terms of the expected Eaves et al.
ACCESSION UNIGENE DESCRIPTION MEAN FOLD- t t CONF. AI CHANGE MAPPING NO. ID AI CHANGE (set 1) (set 2) LEVEL LEVEL IN DATA (NOD-like) (Idd5/NOD) B10.Idd3 THYMUS [Chr, cM] MYELOID/ERYTHROCYTE PRECURSOR L00919 Mm.30038 Erythrocyte protein band 4.1 420 1.18 2.61 3.67 <0.01 NOD 4, 65.7 AA267758 Mm.24043 Erythroblast membrane-associated protein 252 1.37 3.95 2.61 <0.01 number of times a given change would have been expected to occur by chance amongst all ~20,000 probe sets examined. They are given only as a guide to aid the reader in assessing which differences are most likely to be genuine. Eaves et al. Supplemental Table 2A Genes that exhibited a “suggestive” difference in expression in the spleen between NOD-derived strains carrying the Idd5 resistance allele (Idd5 and Idd3/5) and those carrying the susceptible allele (NOD, Idd3 and Idd9). See legend of supplemental Table 1 for a full explanation of the data. ACCESSION UNIGENE DESCRIPTION MEAN FOLD- t t CONF. AI CHANGE MAPPING NO. ID AI CHANGE (set 1) (set 2) LEVEL LEVEL IN DATA (NOD-like) (Idd5/NOD) B10-like SPLEEN [Chr, cM] CELL SURFACE ANTIGEN Z16078 Mm.2692 CD53 antigen 411 -1.17 -3.45 -2.92 <0.5 NOD 3, 50.5 MISCELLANEOUS/UNKNOWN W50102† Homologous to sodium channel b-1 subunit precursor 468 1.22 5.7 4.80 <0.001 >Idd5 AA270493 Mm.30682 ESTs, Highly similar to human deoxyhypusine synthase 171 1.24 2.67 2.96 <0.5 NOD AA013766 Mm.133088 ESTs, Weakly similar to TBX2_mouse TBX2 protein 640 1.12 3.17 2.97 <0.05 AA276909 Mm.102541 ESTs, Weakly similar to Grb10 140 1.31 2.91 2.89 <0.1 AI227079 Mm.22227 ESTs, 421 1.14 2.82 3.38 <0.05 AI585706 114 1.40 3.94 2.78 <0.01 AA561284 282 1.14 3.13 2.68 <0.5 AA013504 Mm.12616 Protein kinase IFN inducible dsRNA dependent inhibitor 135 -1.86 -5.06 -4.30 <0.01 NOD L20315 Mm.3999 Macrophage expressed 1 230 -1.22 -3.81 -2.71 <0.5 X90875 Mm.4038 Fragile X mental retardation gene, autosomal homolog 295 -1.23 -4.21 -2.34 <0.5 NOD AA003646 Mm.29655 ESTs, Highly similar to Signal recognition particle 68 kD 216 -1.34 -4.10 -2.88 <0.1 NOD AA177440 Mm.36756 ESTs 331 -1.16 -3.56 -2.57 <0.5 NOD Supplemental Table 2B Genes that exhibited a “suggestive” difference in expression in the thymus between NOD-derived strains carrying the Idd5 resistance allele (Idd5 and Idd3/5) and those carrying the susceptible allele (NOD, Idd3 and Idd9). Where a ‘’ is shown the two B10-derived strains gave conflicting data. For a full explanation of the data see the legend of supplemental Table 1. †Sequence now withdrawn at submitters request. Eaves et al. ACCESSION UNIGENE DESCRIPTION MEAN FOLD- t t CONF. AI CHANGE MAPPING MAPS NO. ID AI CHANGE (set 1) (set 2) LEVEL LEVEL IN DATA TO (NOD-like) (Idd3-5/NOD) B10.Idd3 THYMUS [Chr., cM] INTERVAL CYTOKINE/RECEPTOR AB000803 Mm.1401 Chemokine (C-X-C) receptor 4 (Cmkar4) 334 2.81 11.17 13.99 <0.001 Idd3/5 1, 67.4 SIGNALLING (INTRACELLULAR) AA120636 Mm.28761 Serine/threonine kinase 25 (Stk25) 106 1.75 1.59 4.25 <0.05 Idd3/5 1, 58.4 MYELOID/ERYTHROCYTE PRECURSOR AA051488 Mm.12961 Rhesus blood group-associated A glycoprotein 480 1.23 1.85 3.49 <0.05 Supplemental Table 3A Genes that exhibited a “suggestive” difference in expression in the spleen between the Idd3/5 double congenic and those strains carrying susceptible alleles at Idd3 and/or Idd5 (NOD, Idd3, Idd5 and Idd9). Tick marks are shown to indicate whether any genes were known to map to the Idd3 or Idd5 congenic intervals themselves. See legend of supplemental Table 1 for a full explanation of the data. Eaves et al. Supplemental Table 3B ACCESSION UNIGENE DESCRIPTION MEAN FOLD- t t CONF. AI CHANGE MAPPING MAPS NO. ID AI CHANGE (set 1) (set 2) LEVEL LEVEL IN DATA TO (NOD-like) (Idd3-5/NOD) B10-like SPLEEN [Chr., cM] INTERVAL CYTOKINE/RECEPTOR AB000803 Mm.1401 Chemokine (C-X-C) receptor 4 (Cmkar4) 426 4.62 25.93 38.24 <0.001 Idd3/5 1, 67.4 AA067236 Mm.3157 Fibroblast growth factor receptor 1 233 1.26 1.81 3.43 <0.5 8, 10.0 SIGNALLING (INTRACELLULAR) AA049327 Mm.28761 Serine/threonine kinase 25 (Stk25) 53 3.98 5.50 6.69 <0.001 Idd3/5 1, 58.4 AA000451 Mm.4284 Guanine nucleotide binding protein, alpha inhibiting 2 69 1.86 2.57 4.54 <0.01 9, 59.0 AA517227 Mm.2995 Ect2 oncogene 197 2.29 2.35 7.65 <0.01 3, B MYELOID/ERYTHROCYTE PRECURSOR AI481796 Mm.35829 Erythroid differentiation regulator 705 1.24 2.05 4.27 <0.05 MISCELLANEOUS/UNKNOWN AA000144 Mm.16831 Creatine kinase, brain 500 1.34 2.65 10.69 <0.01 12, 55.0 AA014646 Mm.2375 Creatine kinase, muscle 242 1.21 2.08 2.51 <0.5 7, 4.5 AA000365 Mm.2580 Syndecan 1 137 1.67 2.56 3.76 <0.05 12, 1.0 AA000631 Mm.2469 Split hand/foot deleted gene 1 276 1.19 2.12 3.58 <0.05 6, A2 AA015381 Mm.12155 Syntaxin binding protein 3 366 1.33 2.72 6.09 <0.01 AA048346 Mm.2006 Transgelin 417 1.27 2.45 3.73 <0.05 9, 27.0 AA098549 Mm.2423 Procollagen, type II, alpha 1 398 1.21 2.17 4.31 <0.05 15, 54.5 AA510381 Mm.25198 Phosphoribosyl pyrophosphate synthetase 1 477 1.16 2.24 4.77 <0.01 X, 60.0 W14596 Mm.14191 Kallikrein binding protein 91 2.60 3.19 3.81 <0.01 AA000067 Mm.3381 Ribosomal protein S8 608 1.12 2.11 3.13 <0.1 AI314508 Mm.42193 Spindlin 184 1.28 1.94 3.27 <0.5 U14103 Mm.4485 Ral guanine nucleotide dissociation stimulator,-like 1 799 1.13 2.16 2.47 <0.5 U71206 Mm.4832 Paired-like homeodomain transcription factor 1 241 1.22 2.47 2.45 <0.5 13, 34.0 AI591846 Mm.20925 G1 to phase transition 1 61 1.57 2.05 2.32 <0.5 16, 3.8 AA422489 Mm.142740 Hypothetical protein MNCb-2146 637 1.26 2.08 4.51 <0.05 AA611734 Mm.142508 DNA segment, Chr 4, ERATO Doi 20, expressed 179 1.70 2.72 3.65 <0.05 4, 51.8 AA497978 Mm.2121 DNA segment, Chr 12, ERATO Doi 647, expressed 943 1.23 2.23 4.98 <0.01 12, 51.0 AA016659 Mm.42275 DNA segment, Chr 9, ERATO Doi 25, expressed 331 1.41 3.07 6.95 <0.001 9, 71.0 AA024189 Mm.29337 Mus musculus brain cDNA, clone MNCb-1820 70 1.48 1.81 2.42 <1 AA840137 Mm.28959 ESTs, Highly similar to antigen NY-CO-1 [H.sapiens] 315 1.15 2.13 2.70 <0.5 NOD AA617093 Mm.29571 ESTs, Highly similar to SEC14L [H.sapiens] 374 1.13 1.89 2.55 <0.5 AA013851 Mm.29972 ESTs, Highly similar to RAP1 [H.sapiens] 430 1.12 2.05 2.80 <0.5 AA140311 Mm.5612 ESTs, Highly similar to eIF 4B [Homo sapiens] 351 1.24 1.86 5.30 <0.05 AA163617 Mm.27421 ESTs, Moderately similar to unnamed protein product 87 1.54 2.08 2.54 <0.5 AA060478 Mm.36726 ESTs, Moderately similar to ladinin [H.sapiens] 318 1.16 2.73 2.34 <0.5 Idd3/5 Eaves et al. (continued) ACCESSION UNIGENE DESCRIPTION MEAN FOLD- t t CONF. AI CHANGE MAPPING MAPS NO. ID AI CHANGE (set 1) (set 2) LEVEL LEVEL IN DATA TO (NOD-like) (Idd3-5/NOD) B10-like SPLEEN [Chr., cM] INTERVAL MISCELLANEOUS/UNKNOWN (continued) D18973 Mm.3069 ESTs, Weakly similar to RIN1 [R.norvegicus] 108 1.47 2.14 2.38 <0.5 AA276060 Mm.36852 ESTs, Weakly similar to AF188634_1 F protein 133 1.32 1.99 2.30 <0.5 AA014952 Mm.41128 ESTs, Weakly similar to zinc finger protein 95 54 1.83 3.04 3.45 <0.01 AA079918 Mm.142315 ESTs, Weakly similar to Unknown 496 1.20 2.86 3.74 <0.01 AA073709 Mm.20207 ESTs 241 1.38 2.68 3.94 <0.05 AA204020 Mm.2085 EST 528 1.51 2.06 6.70 <0.01 AA024106 Mm.44885 ESTs 167 1.27 1.86 3.07 <0.5 AA882052 cDNA clone IMAGE:1277236 75 1.57 1.87 2.91 <0.5 Idd3/5 W10776 cDNA clone IMAGE:314509 73 2.00 2.09 5.07 <0.01 W12894 cDNA clone IMAGE:329872 353 1.32 2.61 3.70 <0.05 AI587983 50 1.72 2.63 2.58 <0.1 AA013663 201 1.22 2.17 2.52 <0.5 W55634 229 1.28 2.92 2.84 <0.05 U25739 62 1.46 1.89 2.32 <1 AA030081 122 1.31 1.96 2.40 <0.5 AI158490 413 1.15 2.11 2.96 <0.1 AA038168 Mm.300 Carbonic anhydrase 3 208 -2.09 -3.77 -3.96 <0.5 NOD 3, 11.7 AI020741 Mm.38421 ESTs 239 -1.32 -3.73 -3.81 <0.5 NOD Supplemental Table 3B Genes that exhibited a “suggestive” difference in expression in the thymus between the Idd3/5 double congenic and those strains carrying susceptible alleles at Idd3 and/or Idd5 (NOD, Idd3, Idd5 and Idd9). A cross indicates that the expression in the B10.Idd3 set 1 sample was higher than in the B10.H2g7 sample. Tick marks are shown to indicate whether any genes were known to map to the Idd3 or Idd5 congenic intervals themselves. See legend of supplemental Table 1 for a full explanation of the data. Eaves et al. ACCESSION UNIGENE DESCRIPTION MEAN FOLD- t t CONF. AI CHANGE MAPPING MAPS NO. ID AI CHANGE (set 1) (set 2) LEVEL LEVEL IN DATA TO (NOD-like) (Idd9/NOD) B10.Idd THYMUS [Chr., cM] INTERVAL 3 MYELOID/ERYTHROCYTE PRECURSOR AA222297 Mm.18140 Rhesus blood group-like 130 Absent -12.28 -7.38 <0.01 Idd9 4, 65.7 C77974 Mm.18140 Rhesus blood group-like 254 Absent -18.72 -9.94 <0.001 Idd9 4, 65.7 C77974 Mm.18140 Rhesus blood group-like 191 -2.52 -6.91 -5.12 <0.05 Idd9 4, 65.7 MISCELLANEOUS/UNKNOWN AA000144 Mm.16831 Creatine kinase, brain 504 1.15 2.67 2.64 <0.5 Idd9 12, 55.0 AA108699 Mm.10812 Phosphodiesterase 9A 834 1.26 3.55 2.32 <0.5 Idd9 17 U88327 Mm.4132 Cytokine inducible SH2-containing protein 2 211 1.25 2.37 2.39 <0.5 Idd9 10, 52.0 AI098764 Mm.30063 Ribonuclease, RNase A family 4 98 1.65 3.68 2.09 <0.5 NOD AA422489 Mm.142740 Hypothetical protein MNCb-2146 552 1.16 3.16 2.26 <0.5 NOD 8 W29408 Mm.28357 DNA segment, Chr 16, Wayne State University 103 308 1.18 2.41 2.22 <0.5 NOD 16, 9.9 AA182335 Mm.76675 ESTs 532 1.16 2.59 2.16 <1 NOD AA002308 Mm.83840 ESTs 246 1.38 3.95 2.83 <0.05 NOD AA204020 Mm.2085 EST 506 1.37 4.06 2.49 <0.1 NOD AA529269 Mm.911 High mobility group protein 17 339 -1.69 -3.95 -6.33 <0.5 Idd9 4, 62.2 AA185951 Mm.43213 DNA segment, Chr 4, ERATO Doi 314, expressed 130 -4.74 -4.62 -5.08 <0.5 Idd9 4, 57.6 AA009152 Mm.88349 ESTs, Highly similar to small membrane protein 1 359 -1.54 -3.96 -7.22 <0.5 Idd9 4, ~65* AA032327 Mm.21873 ESTs, Weakly similar to predicted [C.elegans] 211 -1.60 -4.30 -4.40 <0.5 Idd9 8 AA097418 Mm.4554 Lysosomal-associated protein transmembrane 5 202 -2.07 -4.12 -6.62 <0.5 NOD 4, ~60* Supplemental Table 4A Genes that exhibited a “suggestive” difference in expression in the spleen between the Idd9 strain and those carrying the NOD susceptibility allele at Idd9 (NOD, Idd3, Idd5 and Idd3/5). Tick marks are shown to indicate whether any genes were known to map to the Idd9R28 congenic interval itself. Genes that had an AI of less than 20 are shown as absent, rather than specifying a particular fold-change. See legend of supplemental Table 1 for a full explanation of the data. *Approximate position inferred from human orthologue. Eaves et al. ACCESSION UNIGENE DESCRIPTION MEAN FOLD- t t CONF. AI CHANGE MAPPING MAPS NO. ID AI CHANGE (set 1) (set 2) LEVEL LEVEL IN DATA TO (NOD-like) (Idd9/NOD) B10.Idd3 SPLEEN [Chr., cM] INTERVAL CELL SURFACE/RECEPTOR U07634† Mm.2581 Eph receptor A2 209 -1.91 -3.72 -11.80 <0.05 Idd9 4, 73.2 U21795† Mm.2923 Interleukin 2 receptor, gamma chain 136 -1.90 -3.10 -5.50 <1 NOD X, 38.0 MISCELLANEOUS/UNKNOWN W29229 Mm.27663 Transcription elongation factor B (SIII), polypeptide 3 139 1.45 2.75 2.61 <0.1 Idd9 4, D3 U63419 Mm.4177 Uncoupling protein, mitochondrial 194 1.75 5.87 5.37 <0.001 NOD 8, 38.0 AI118299 Mm.21417 Apolipoprotein E 521 1.21 2.24 2.31 <0.5 x AA276791 Mm.22699 Selenoprotein P, plasma 1 369 1.14 2.25 4.84 <0.01 >Idd9 15, 5.9 AA175675 Mm.100378 ESTs, Highly similar to ENV polyprotein precursor 176 1.47 2.39 4.86 <0.01 NOD AA475531 Mm.21580 ESTs; Highly similar to -L-fucosidase precursor [Rn] 214 1.21 2.48 2.45 <0.5 NOD AA267055 Mm.22176 ESTs 70 2.04 2.77 4.14 <0.01 Idd9 4, 65.7 AA529269† Mm.911 High mobility group protein 17 818 -1.24 -2.98 -4.05 2.5 Idd9 4, 62.2 AA097418 Mm.4554 Lysosomal-associated protein transmembrane 5 297 -2.13 -9.56 -14.50 <0.001 NOD 4, ~60* AA009152 Mm.88349 ESTs, Highly similar to small membrane protein 1 413 -1.60 -5.41 -11.16 <0.01 Idd9 4, ~65* AA008202 Mm.22409 Chloride intracellular channel 4 (mitochondrial) 414 -1.43 -4.60 -8.81 <0.05 NOD AA422356† Mm.22409 Chloride intracellular channel 4 (mitochondrial) 250 -1.61 -2.89 -9.78 <0.05 NOD AA501061† Mm.33120 ESTs, Highly similar to rat histamine n-methyltransferase 81 -2.54 -2.60 -7.32 <0.5 NOD 2 AA238723 Mm.46248 ESTs, Highly similar to rat peptide YY precursor 458 -1.36 -7.37 -5.88 <0.1 NOD AA000215† Mm.46777 ESTs, Weakly similar to Ylr239cp [S.cerevisiae] 141 -1.81 -4.25 -5.41 <0.5 NOD Supplemental Table 4B Genes that exhibited a “suggestive” difference in expression in the thymus between the Idd9 strain and those carrying the NOD susceptibility allele at Idd9 (NOD, Idd3, Idd5 and Idd3/5). Tick marks are shown to indicate whether any genes were known to map to the Idd9R28 congenic interval itself. See legend of supplemental Table 1 for a full explanation of the data. †Probe set did not meet criteria for a suggestive change using original thresholds (see text for explanation). *Approximate position inferred from human orthologue. Eaves et al. ACCESSION UNIGENE DESCRIPTION MEAN FOLD- t t CONF. AI CHANGE MAPPING NO. ID AI CHANGE (set 1) (set 2) LEVEL LEVEL IN 2 DATA (Congenics) (NOD/Cong.) B10.Idd3 TISSUE [Chr., cM] SPLEEN X03151 Mm.3951 Thymus cell antigen 1; theta (CD90) 379 1.19 1.97 3.55 <0.01 NOD 9, 26.0 AA673702 cDNA clone IMAGE:1150049 5', mRNA sequence 213 1.29 1.76 2.4 <0.5 NOD THYMUS D85570 Proteasome (prosome, macropain) subunit, beta type 7 303 1.21 2.41 1.86 <1 NOD 2, 27.5 W10127 Mm.16763 Aldolase 1, A isoform 666 1.39 2.48 2.48 <0.1 congenic AI390779 cDNA clone IMAGE:654065 84 1.49 2.55 1.89 <0.5 congenic AA049063 cDNA clone IMAGE:479564 5', mRNA sequence 265 -1.54 -6.20 -4.87 <0.05 congenic AA498580 cDNA clone IMAGE:889874 5', mRNA sequence 86 Absent -3.50 -16.32 <0.01 congenic Supplemental Table 5 Genes that exhibited a “suggestive” difference in expression between the parental NOD strain and diabetes resistant NOD- derived congenic strains (Idd3, Idd5, Idd3/5 and Idd9). Genes that had an AI of less than 20 are shown as absent, rather than specifying a particular fold- change. See legend of supplemental Table 1 for a full explanation of the data.