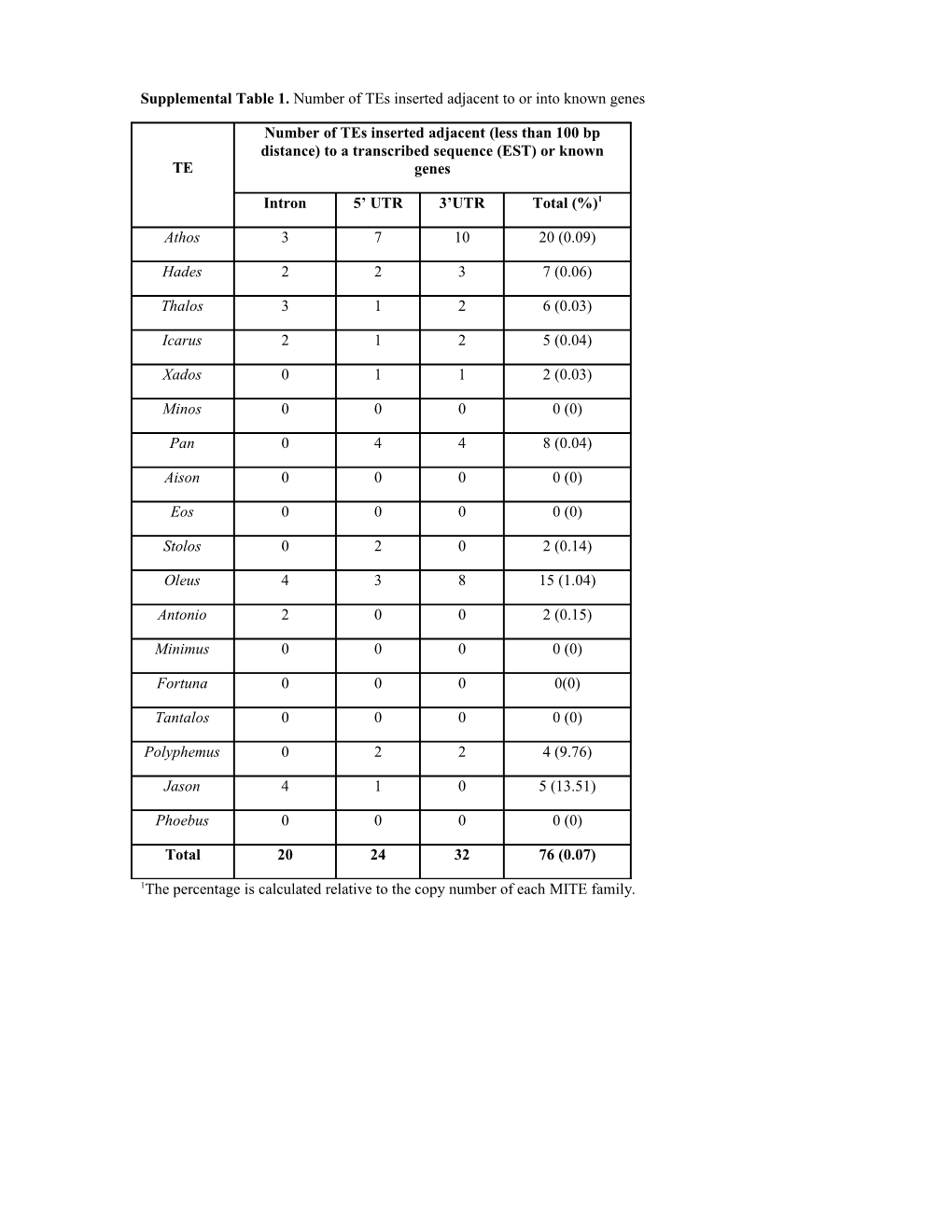
Supplemental Table 1. Number of TEs inserted adjacent to or into known genes
Number of TEs inserted adjacent (less than 100 bp distance) to a transcribed sequence (EST) or known TE genes
Intron 5’ UTR 3’UTR Total (%)1
Athos 3 7 10 20 (0.09)
Hades 2 2 3 7 (0.06)
Thalos 3 1 2 6 (0.03)
Icarus 2 1 2 5 (0.04)
Xados 0 1 1 2 (0.03)
Minos 0 0 0 0 (0)
Pan 0 4 4 8 (0.04)
Aison 0 0 0 0 (0)
Eos 0 0 0 0 (0)
Stolos 0 2 0 2 (0.14)
Oleus 4 3 8 15 (1.04)
Antonio 2 0 0 2 (0.15)
Minimus 0 0 0 0 (0)
Fortuna 0 0 0 0(0)
Tantalos 0 0 0 0 (0)
Polyphemus 0 2 2 4 (9.76)
Jason 4 1 0 5 (13.51)
Phoebus 0 0 0 0 (0)
Total 20 24 32 76 (0.07)
1The percentage is calculated relative to the copy number of each MITE family. Supplemental Table 2. List of MITE-flanking genes and their distances from the insertion (in bp), upstream of the 5’UTR, or downstream of the 3’UTR of the gene.
Gene annotation TE insertion Accession number Distance from gene (bp) cell division protein AAA ATPase family Athos EU835980.1 305 upstream 5K14.8 Athos AF488415.1 205 upstream VatpC Athos EU358770.2 573 upstream VatpC Athos EU358770.2 653 upstream TAK19-1 Athos FJ447462.1 31 upstream CD892187_2 Athos EU358770.2 355 upstream Acc-1 Athos EU660902.1 982 downstream CBF12 Athos AY951944.1 581 downstream Acc-1 Athos EU660897.1 982 downstream AP2-L Athos EU358770.2 364 downstream Cf2.2 Athos EU358770.2 419 downstream Cf2.1 Athos EU358770.2 436 downstream 2050O8.4 Athos GQ165812.1 731 downstream Cf2.1 Athos EU358770.2 394 downstream Cf2.2 Athos EU358770.2 377 downstream putative receptor kinase Hades AY494981.1 512 upstream disease resistance protein (LR1) Hades EF567062.1 457 upstream cell division AAA ATPase family protein Hades EU835981.1 474 downstream cytochrome P450 Hades FJ436983.1 268 downstream hypothetical protein Hades GU817319.1 962 downstream chloroplast pseudogene encoding ATP synthase CFO Icarus AY951945.1 410 upstream subunit I single strand DNA repair-like protein Icarus AY951945.1 938 downstream putative polyprotein Icarus FJ447462.1 895 downstream unknown Jason AY491681.1 288 upstream Tm_AY485644.9 Oleus AY485644.1 926 upstream ACT-1 Oleus AY663392.1 569 upstream ACT-1 Oleus AY663391.1 577 upstream WM1.3 Oleus AY534123.1 314 downstream putative receptor kinase Oleus AY494981.1 217 downstream receptor kinase 2 Oleus DQ537335.1 87 downstream CBF2 Oleus AY951945.1 274 downstream WM1.3 Oleus AY534123.1 259 downstream FtsH4 Oleus EU358773.2 401 downstream LMW glutenin precursor Oleus EF426565.1 405 downstream FtsH4 Oleus EU358773.2 279 downstream putative receptor kinase Pan AY494981.1 759 upstream putative receptor kinase Pan AY494981.1 697 upstream receptor kinase 2 Pan DQ537335.1 758 upstream receptor kinase 2 Pan DQ537335.1 696 upstream putative receptor kinase Pan AY494981.1 352 downstream receptor kinase 2 Pan DQ537335.1 222 downstream Acc-1 Pan EU660901.1 596 downstream Acc-1 Pan EU660899.1 596 downstream putative ABC transporter Polyphemus AF459639.1 652 upstream P450 Polyphemus EF567062.1 706 upstream ABP-1 Polyphemus EF567062.1 679 downstream hypothetical protein Polyphemus GU817319.1 798 downstream putative receptor kinase Stolos AY494981.1 189 upstream ZCCT1 Stolos EF540321.1 753 upstream CBF9 Thalos AY951945.1 632 upstream Acc-1 Thalos EU660902.1 633 downstream Acc-1 Thalos EU660897.1 633 downstream unknown Xados EU835980.1 239 upstream putative PEX11-1 protein Xados GQ165812.1 299 downstream Supplemental Table 3. List of wheat species and accessions analyzed in this study and their geographic origin.
Genus Species Accession1 Country Region 599124 Israel West Bank 599140 Jordan NA 599149 Israel Southern 599173 Israel Southern Aegilops searsii TE16 Syria Gabagib TE36 Syria NA TE44 Jordan Shonbak AEG-1589 Israel Southern Clae45 Unknown NA 542274 Turkey Adiyaman TS01 Israel Berekhya TS02 Israel Zikhron Ya'akov Aegilops speltoides TS118 Syria NA TS41 Israel Wadi Hilazon TS46 Turkey Izmir TS47 Unknown NA 6008 Israel NA TQ27 NA NA 2001 Pakistan Balochistan 268210 Iran Mazandaran 369627 Unknown NA 452131 China Qinghai Aegilops tauschii 574468 Arminia NA 603236 Turkmenistan NA 603237 Azerbaijan NA 603246 Portugal NA KU2069 Iran Tehran TMU38 NA NA TMU06 NA NA TMU15 NA NA Triticum urartu TMU16 NA NA TMU34 NA NA TMU44 NA NA TMU47 NA NA Triticum monococcum TMB02 NA NA Aegilops sharonensis TH02 Israel Acre Nahariya-Rosh Aegilops longissima TL05 Israel Hanukra road Triticum dicoccoides TTD20 Israel NA Triticum durum TTR19 NA NA Triticum aestivum TAA01 NA NA 1 Accessions refer to Plant IDs or USDA inventory numbers. NA, data not available. Supplemental Table 4. List of primers used for quantitative PCR.
TE Forward primer Reverse primer Efficiency1 Fortuna GGATGCGGGAGTGAGAAGG CTCCCATGCACAACAAGCC 100% Minos GTAGTGCTTTCTCTATCCACGTGC TTTACTTTCGCCGCAATCG 91% Oleus CTCCCTCCGTTCACTATTATAAGATGT CATACGGACTGAAATGAGTGAACAA 100% Eos GGGTTTGGAGGATAATAAATGCTC ATGCTAGCCTCACCCATGCTT 100% Aison CTACCTCCGTCTCGGTGAATAAGT AATCGTCAACCTAGAACTACGCG 100% Icarus CCGTCCGGAAATACTTGTCG TGTCGCAAAATTAAAATGGATGTATC 98.8% Phoebus CAAAATTAATCTCGGCTAAATCATTG TTGCTCCTGATTGCATGTATCTCT 100% Polyphemus TGTTAACTGCGTGTCACCAAAAA CATAATGTGCATGCCACCAAA 98.5% Stolos CCCTCCGGTCATTTTTAGTCTG TGGCATGGTGAATGTTGATAATTT 95% 1 See Materials and Methods for details on calculating primer efficiency. Supplemental Table 5. List of TE-specific primers used for TMD reactions.
TE Primer sequence Aison TTAAATCGTCAACCTAGAACTACGC Antonio TTTGGGTGTCGTCTTAAGTGTC Eos CAGGGGTGCTTGGAACTTTA Fortuna GTGGCCTTATATAGCTGCAAA Hades GTACTAAGCTTGAGACACTTATTTTGG Jason TTGACTTAGGACAAACTTAGAACTGC Minos GTCAAAATTGAAGCACGTGGA Oleus CAAATCAGAAAGCTGGAACATC Phoebus TGATCACCGTGTCAAAAATAATACA Polyphemus CATAAAATACGAAGGGGCACA Tantalos TCTAGACAAACCTAGTATGCGGAGT Thalos GCTCCGTATGTAGTCACTTATTGA Xados TCTGAAACAAGTATTTCCGAACG