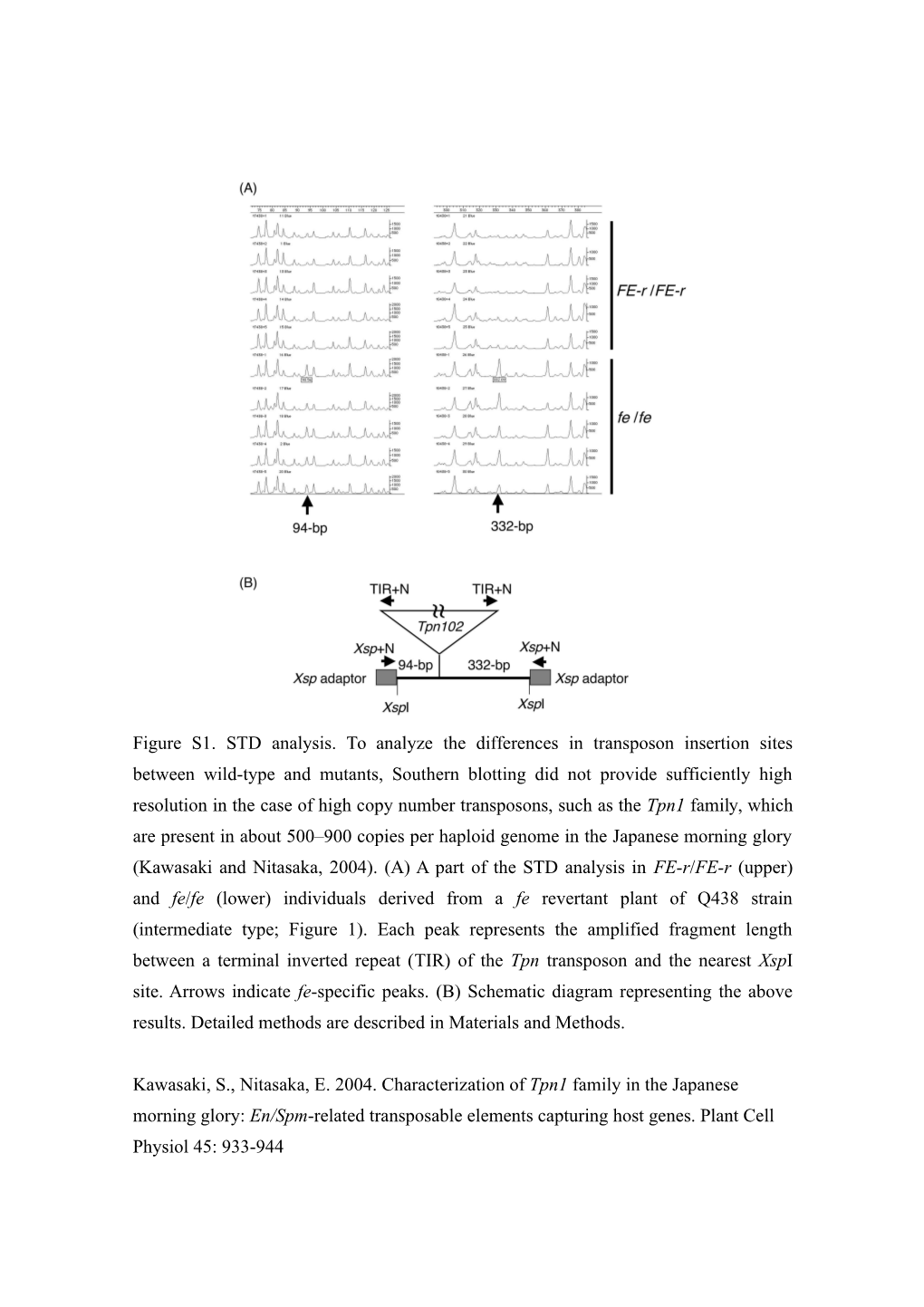
Figure S1. STD analysis. To analyze the differences in transposon insertion sites between wild-type and mutants, Southern blotting did not provide sufficiently high resolution in the case of high copy number transposons, such as the Tpn1 family, which are present in about 500–900 copies per haploid genome in the Japanese morning glory (Kawasaki and Nitasaka, 2004). (A) A part of the STD analysis in FE-r/FE-r (upper) and fe/fe (lower) individuals derived from a fe revertant plant of Q438 strain (intermediate type; Figure 1). Each peak represents the amplified fragment length between a terminal inverted repeat (TIR) of the Tpn transposon and the nearest XspI site. Arrows indicate fe-specific peaks. (B) Schematic diagram representing the above results. Detailed methods are described in Materials and Methods.
Kawasaki, S., Nitasaka, E. 2004. Characterization of Tpn1 family in the Japanese morning glory: En/Spm-related transposable elements capturing host genes. Plant Cell Physiol 45: 933-944