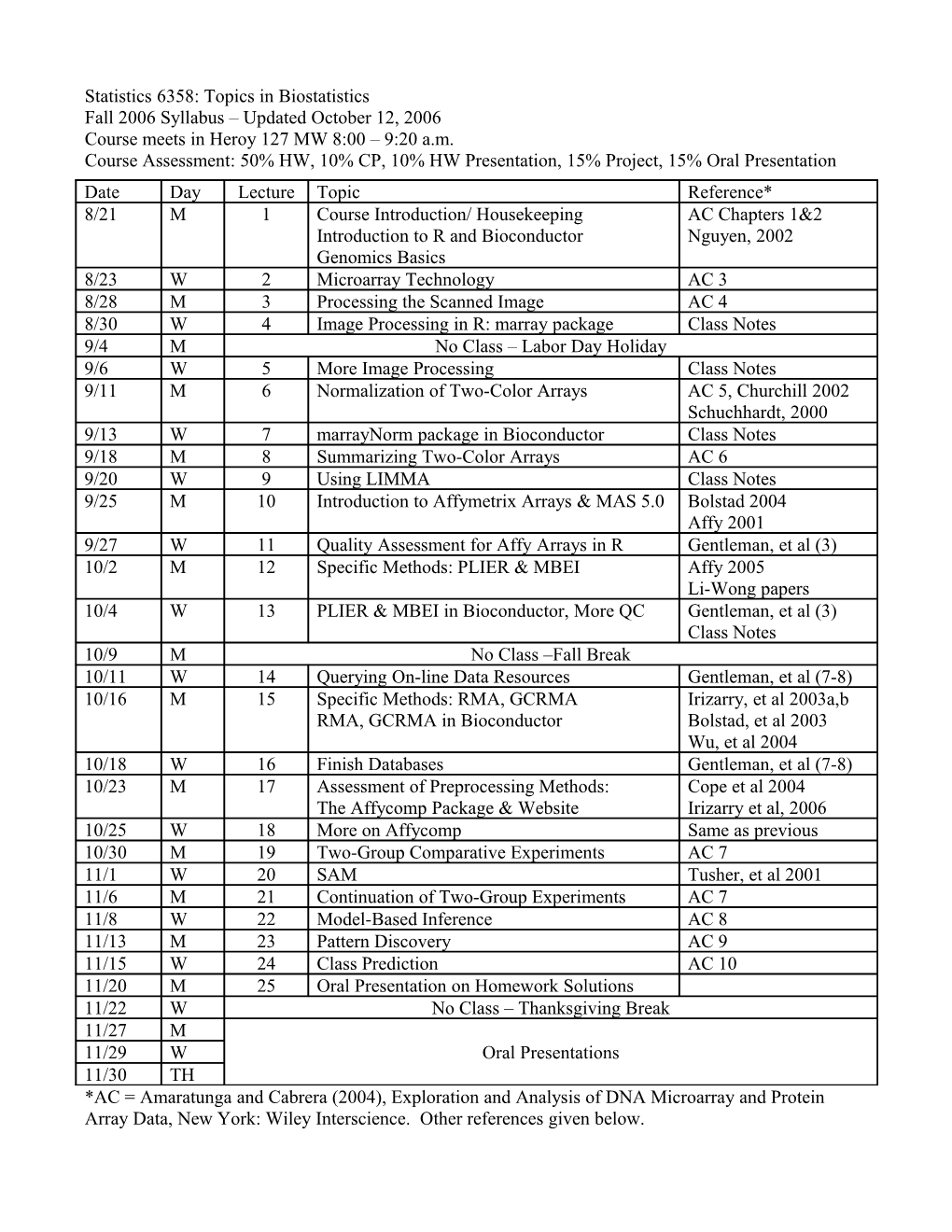
Statistics 6358: Topics in Biostatistics Fall 2006 Syllabus – Updated October 12, 2006 Course meets in Heroy 127 MW 8:00 – 9:20 a.m. Course Assessment: 50% HW, 10% CP, 10% HW Presentation, 15% Project, 15% Oral Presentation Date Day Lecture Topic Reference* 8/21 M 1 Course Introduction/ Housekeeping AC Chapters 1&2 Introduction to R and Bioconductor Nguyen, 2002 Genomics Basics 8/23 W 2 Microarray Technology AC 3 8/28 M 3 Processing the Scanned Image AC 4 8/30 W 4 Image Processing in R: marray package Class Notes 9/4 M No Class – Labor Day Holiday 9/6 W 5 More Image Processing Class Notes 9/11 M 6 Normalization of Two-Color Arrays AC 5, Churchill 2002 Schuchhardt, 2000 9/13 W 7 marrayNorm package in Bioconductor Class Notes 9/18 M 8 Summarizing Two-Color Arrays AC 6 9/20 W 9 Using LIMMA Class Notes 9/25 M 10 Introduction to Affymetrix Arrays & MAS 5.0 Bolstad 2004 Affy 2001 9/27 W 11 Quality Assessment for Affy Arrays in R Gentleman, et al (3) 10/2 M 12 Specific Methods: PLIER & MBEI Affy 2005 Li-Wong papers 10/4 W 13 PLIER & MBEI in Bioconductor, More QC Gentleman, et al (3) Class Notes 10/9 M No Class –Fall Break 10/11 W 14 Querying On-line Data Resources Gentleman, et al (7-8) 10/16 M 15 Specific Methods: RMA, GCRMA Irizarry, et al 2003a,b RMA, GCRMA in Bioconductor Bolstad, et al 2003 Wu, et al 2004 10/18 W 16 Finish Databases Gentleman, et al (7-8) 10/23 M 17 Assessment of Preprocessing Methods: Cope et al 2004 The Affycomp Package & Website Irizarry et al, 2006 10/25 W 18 More on Affycomp Same as previous 10/30 M 19 Two-Group Comparative Experiments AC 7 11/1 W 20 SAM Tusher, et al 2001 11/6 M 21 Continuation of Two-Group Experiments AC 7 11/8 W 22 Model-Based Inference AC 8 11/13 M 23 Pattern Discovery AC 9 11/15 W 24 Class Prediction AC 10 11/20 M 25 Oral Presentation on Homework Solutions 11/22 W No Class – Thanksgiving Break 11/27 M 11/29 W Oral Presentations 11/30 TH *AC = Amaratunga and Cabrera (2004), Exploration and Analysis of DNA Microarray and Protein Array Data, New York: Wiley Interscience. Other references given below. References: Links to references in green are available on the course website (References). Links to the others will require copyright permission, and I am working on it. If you are impatient, you can find them yourself!
Affymetrix, Inc. (2002) Statistical algorithms description document. Affymetrix, Inc. (2005) Technical note: guide to probe logarithmic intensity error (PLIER) estimation. Bolstad BM. (2004) Low Level Analysis of High-density oligonucleotide array data: Background, normalization and summarization [dissertation]. Department of Statistics, University of California at Berkeley, http://bmbolstad.com/ Bolstad, B.M. et al. (2003) A comparison of normalization methods for high density oligunucleotide array data based on variance and bias. Bioinformatics, 19, 185-193 Cui, X., Kerr, M., and Churchill, G (2003) Transformations for cDNA micorarray data, Statistical Methods in Genetics and Molecular Biology, 2(1) article 4 Cope, L.M. et.al. (2003) A benchmark for Affymetrix GeneChip expression measures, Bioinformatics, 20, 323-331. Gentleman, R., Huber, W., Carey, V.J., Irizarry, R.A., Dudoit, S. (eds). Bioinformatics and Computational Biology Solutions Using R and Bioconductor (2005). New York: Springer Irizarry, R.A. et al. (2003a) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res., 31, 1-8 Irizarry,R.A. et al. (2003b) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics, 4, 249-264. Irizarry, R.A. et al. (2006) Comparison of Affymetrix GeneChip expression measures. Bioinformatics, 22, 789-794. Klebanov L. and Yakovlev A., (2006) Treating expression levels of different genes as a sample in microarray data analysis: Is it worth a risk? Statistical Applications in Genetics and Molecular Biology, 5 (1), Article 9. Li, C and Wong, H.W. (2001a) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Nat. Acad. Sci., 98, 31-36. Li, C and Wong, H.W. (2001b) Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol., 2, research0032.1-0032.11. Nguyen, D.V., Arpat, A.B., Wang, N., and Carroll, R.J. (2002) DNA Microarray Experiments: Biological and Technical Aspects, Biometrics 58, 701-717. Schuchhardt, J., Beule,D. et al. (2000) Normalization strategies for cDNA microarrays. Nucleic Acids Research 28, No. 10. Wu, Z. and Irizarry, R.A (2005) A Statistical Framework for the Analysis of Microarray Probe-Level Data, Johns Hopkins University, Dept. of Biostatistics Working Papers. Working Paper 73. http://www.bepress.com/jhubiostat/paper73 Wu, Z. et.al. (2004) A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc., 99, 909-917. Yang, Y.H., Dudoit, S., Luu, P. and Speed, T.P. (2001) Normalization for cDNA microarray data, UC Berkeley Statistics Technical Report: http://www.stat.berkeley.edu/users/terry/zarray/Html/normspie.html