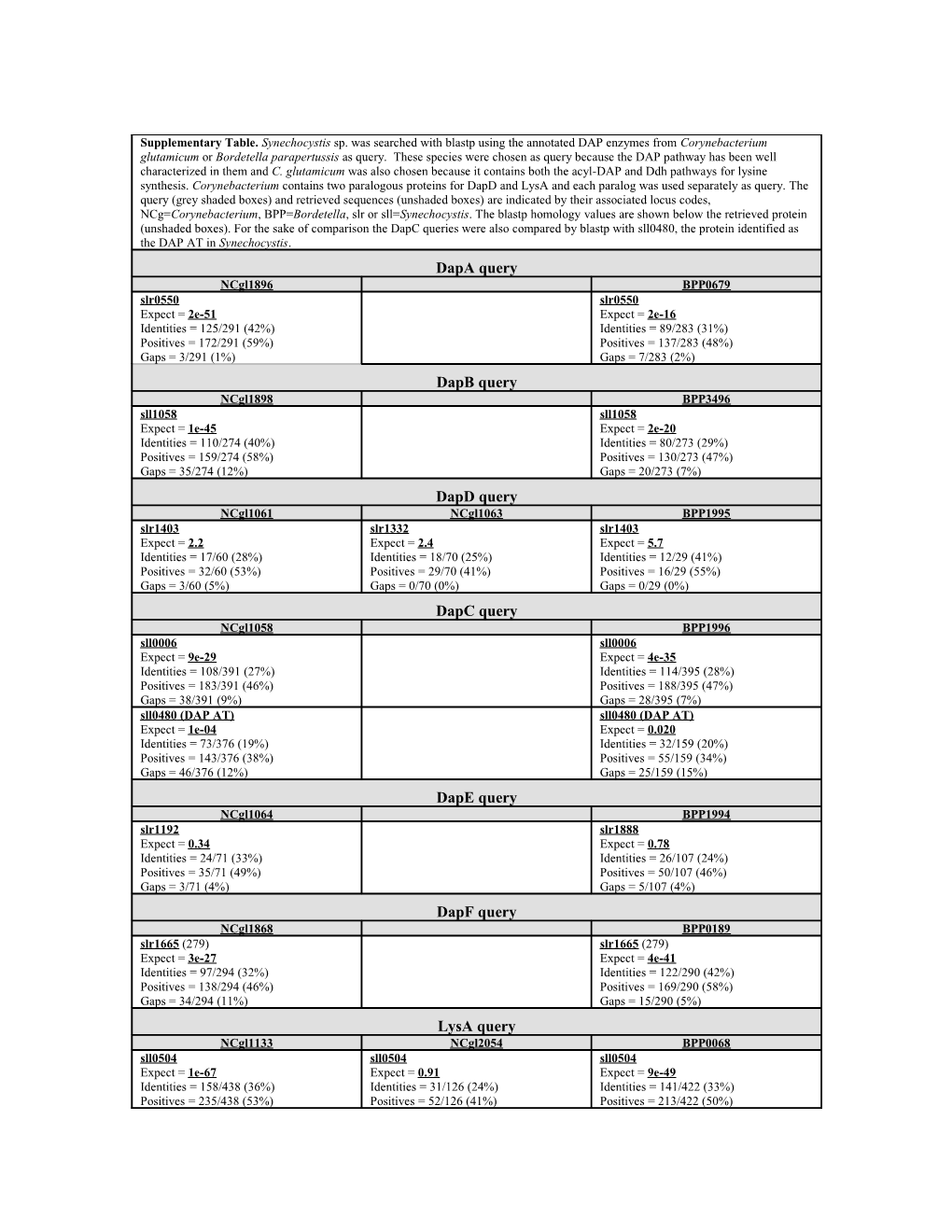
Supplementary Table. Synechocystis sp. was searched with blastp using the annotated DAP enzymes from Corynebacterium glutamicum or Bordetella parapertussis as query. These species were chosen as query because the DAP pathway has been well characterized in them and C. glutamicum was also chosen because it contains both the acyl-DAP and Ddh pathways for lysine synthesis. Corynebacterium contains two paralogous proteins for DapD and LysA and each paralog was used separately as query. The query (grey shaded boxes) and retrieved sequences (unshaded boxes) are indicated by their associated locus codes, NCg=Corynebacterium, BPP=Bordetella, slr or sll=Synechocystis. The blastp homology values are shown below the retrieved protein (unshaded boxes). For the sake of comparison the DapC queries were also compared by blastp with sll0480, the protein identified as the DAP AT in Synechocystis. DapA query NCgl1896 BPP0679 slr0550 slr0550 Expect = 2e-51 Expect = 2e-16 Identities = 125/291 (42%) Identities = 89/283 (31%) Positives = 172/291 (59%) Positives = 137/283 (48%) Gaps = 3/291 (1%) Gaps = 7/283 (2%) DapB query NCgl1898 BPP3496 sll1058 sll1058 Expect = 1e-45 Expect = 2e-20 Identities = 110/274 (40%) Identities = 80/273 (29%) Positives = 159/274 (58%) Positives = 130/273 (47%) Gaps = 35/274 (12%) Gaps = 20/273 (7%) DapD query NCgl1061 NCgl1063 BPP1995 slr1403 slr1332 slr1403 Expect = 2.2 Expect = 2.4 Expect = 5.7 Identities = 17/60 (28%) Identities = 18/70 (25%) Identities = 12/29 (41%) Positives = 32/60 (53%) Positives = 29/70 (41%) Positives = 16/29 (55%) Gaps = 3/60 (5%) Gaps = 0/70 (0%) Gaps = 0/29 (0%) DapC query NCgl1058 BPP1996 sll0006 sll0006 Expect = 9e-29 Expect = 4e-35 Identities = 108/391 (27%) Identities = 114/395 (28%) Positives = 183/391 (46%) Positives = 188/395 (47%) Gaps = 38/391 (9%) Gaps = 28/395 (7%) sll0480 (DAP AT) sll0480 (DAP AT) Expect = 1e-04 Expect = 0.020 Identities = 73/376 (19%) Identities = 32/159 (20%) Positives = 143/376 (38%) Positives = 55/159 (34%) Gaps = 46/376 (12%) Gaps = 25/159 (15%) DapE query NCgl1064 BPP1994 slr1192 slr1888 Expect = 0.34 Expect = 0.78 Identities = 24/71 (33%) Identities = 26/107 (24%) Positives = 35/71 (49%) Positives = 50/107 (46%) Gaps = 3/71 (4%) Gaps = 5/107 (4%) DapF query NCgl1868 BPP0189 slr1665 (279) slr1665 (279) Expect = 3e-27 Expect = 4e-41 Identities = 97/294 (32%) Identities = 122/290 (42%) Positives = 138/294 (46%) Positives = 169/290 (58%) Gaps = 34/294 (11%) Gaps = 15/290 (5%) LysA query NCgl1133 NCgl2054 BPP0068 sll0504 sll0504 sll0504 Expect = 1e-67 Expect = 0.91 Expect = 9e-49 Identities = 158/438 (36%) Identities = 31/126 (24%) Identities = 141/422 (33%) Positives = 235/438 (53%) Positives = 52/126 (41%) Positives = 213/422 (50%) Gaps = 21/438 (4%) Gaps = 9/126 (7%) Gaps = 20/422 (4%)
Ddh query NCgl2528 slr0355 Expect = 0.015 Identities = 14/37 (37%) Positives = 24/37 (64%) Gaps = 0/37 (0%)
The results in the supplementary table show that DapA, DapB, DapF and LysA orthologs can be readily identified in Synechocystis sp. based on homology with Corynebacterium and Bordetella coding sequences for these proteins. DapD and DapE orthologs cannot be identified. The best matches have E scores that are no greater than randomly selected sequences. DapC orthologs can be identified. The best match to Corynebacterium DapC shows an E value of 9e-29 and to Bordetella DapC, 4e-35. The Corynebacterium and Bordetella DapC show much lower homology to the LL-DAP aminotransferase sll0480, and the DapC orthologs in Synechocystis show lower homology than the most divergent LL-DAP aminotransferase orthologs (e.g. sll0480 and At4g33680 show an E value of 7.0e-86). In total, the results suggest that Synechocystis lacks the acyl and Ddh pathways for lysine synthesis and have an LL-DAP aminotransferase pathway.