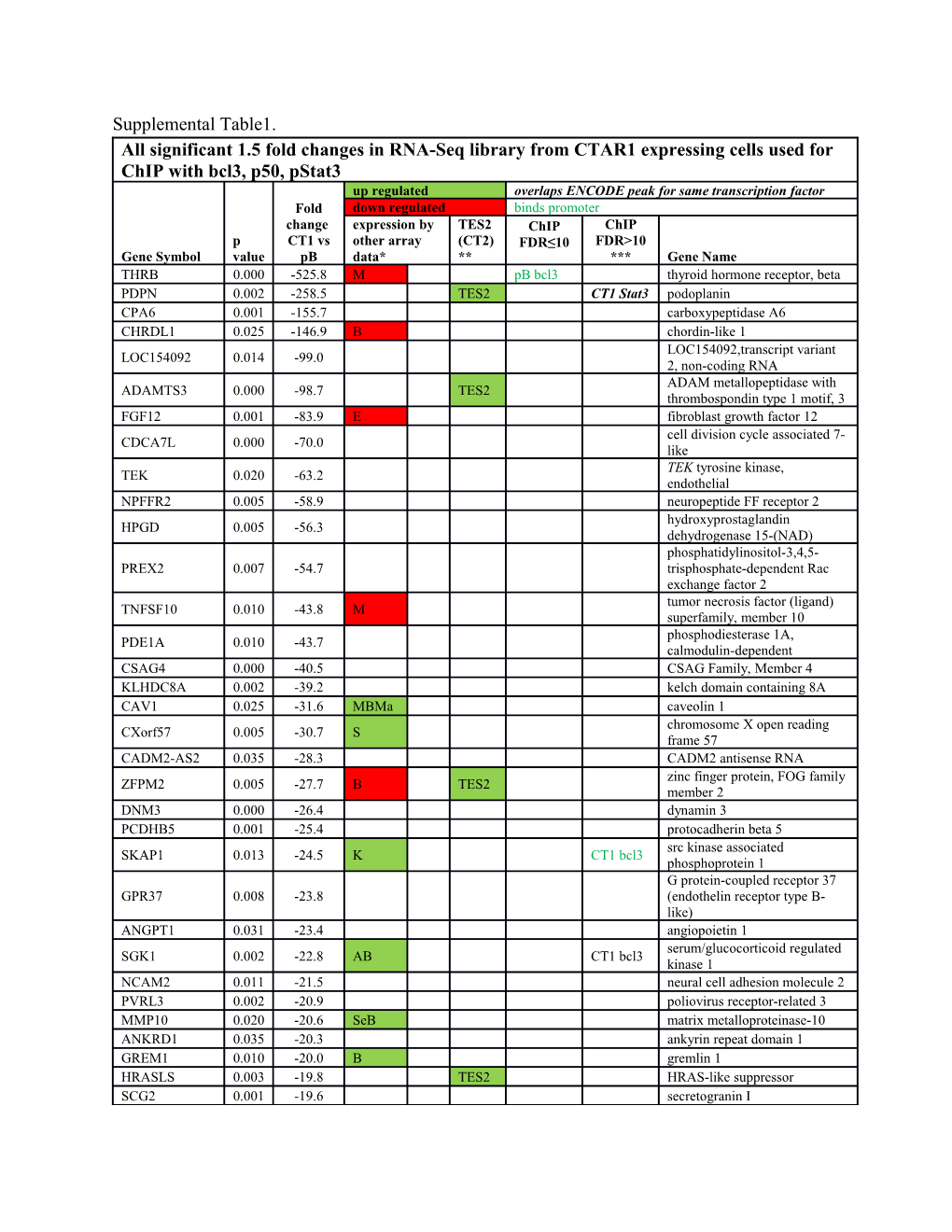
Supplemental Table1. All significant 1.5 fold changes in RNA-Seq library from CTAR1 expressing cells used for ChIP with bcl3, p50, pStat3 up regulated overlaps ENCODE peak for same transcription factor Fold down regulated binds promoter change expression by TES2 ChIP ChIP p CT1 vs other array (CT2) FDR≤10 FDR>10 Gene Symbol value pB data* ** *** Gene Name THRB 0.000 -525.8 M pB bcl3 thyroid hormone receptor, beta PDPN 0.002 -258.5 TES2 CT1 Stat3 podoplanin CPA6 0.001 -155.7 carboxypeptidase A6 CHRDL1 0.025 -146.9 B chordin-like 1 LOC154092,transcript variant LOC154092 0.014 -99.0 2, non-coding RNA ADAM metallopeptidase with ADAMTS3 0.000 -98.7 TES2 thrombospondin type 1 motif, 3 FGF12 0.001 -83.9 E fibroblast growth factor 12 cell division cycle associated 7- CDCA7L 0.000 -70.0 like TEK tyrosine kinase, TEK 0.020 -63.2 endothelial NPFFR2 0.005 -58.9 neuropeptide FF receptor 2 hydroxyprostaglandin HPGD 0.005 -56.3 dehydrogenase 15-(NAD) phosphatidylinositol-3,4,5- PREX2 0.007 -54.7 trisphosphate-dependent Rac exchange factor 2 tumor necrosis factor (ligand) TNFSF10 0.010 -43.8 M superfamily, member 10 phosphodiesterase 1A, PDE1A 0.010 -43.7 calmodulin-dependent CSAG4 0.000 -40.5 CSAG Family, Member 4 KLHDC8A 0.002 -39.2 kelch domain containing 8A CAV1 0.025 -31.6 MBMa caveolin 1 chromosome X open reading CXorf57 0.005 -30.7 S frame 57 CADM2-AS2 0.035 -28.3 CADM2 antisense RNA zinc finger protein, FOG family ZFPM2 0.005 -27.7 B TES2 member 2 DNM3 0.000 -26.4 dynamin 3 PCDHB5 0.001 -25.4 protocadherin beta 5 src kinase associated SKAP1 0.013 -24.5 K CT1 bcl3 phosphoprotein 1 G protein-coupled receptor 37 GPR37 0.008 -23.8 (endothelin receptor type B- like) ANGPT1 0.031 -23.4 angiopoietin 1 serum/glucocorticoid regulated SGK1 0.002 -22.8 AB CT1 bcl3 kinase 1 NCAM2 0.011 -21.5 neural cell adhesion molecule 2 PVRL3 0.002 -20.9 poliovirus receptor-related 3 MMP10 0.020 -20.6 SeB matrix metalloproteinase-10 ANKRD1 0.035 -20.3 ankyrin repeat domain 1 GREM1 0.010 -20.0 B gremlin 1 HRASLS 0.003 -19.8 TES2 HRAS-like suppressor SCG2 0.001 -19.6 secretogranin I WBP5 0.037 -18.4 WW domain binding protein 5 NAA11 0.004 -18.4 N(alpha)-acetyltransferase 11 Neuronal cell adhesion NRCAM 0.005 -17.0 molecule ACTL8 0.011 -16.7 pB bcl3 CT1 Stat3 actin-like 8 PRRG1 0.043 -15.9 A TES2 P53-Responsive Gene 1 chondrosarcoma associated CSAG1 0.008 -14.3 gene 1 DMD 0.001 -14.1 SeB dystrophin platelet-derived growth factor PDGFRA 0.004 -14.0 † CT1 Stat3 bcl3 com receptor, alpha polypeptide S100 calcium binding protein S100A16 0.000 -14.0 A16 RNF128 0.049 -13.9 Se ring finger protein 128 solute carrier family 1 (high SLC1A6 0.005 -13.9 TES2 affinity aspartate/glutamate transporter), member 6 HORMA domain-containing HORMAD1 0.028 -13.4 protein 1 MMP3 0.009 -13.3 B matrix metallopeptidase 3 dickkopf WNT signaling DKK1 0.019 -13.1 B pathway inhibitor 1 GPM6A 0.025 -12.9 M S TES2 glycoprotein M6A RORB 0.018 -12.9 RAR-related orphan receptor B carboxypeptidase, vitellogenic- CPVL 0.001 -12.9 like LEMD1 0.007 -12.7 LEM domain containing 1 SLIT and NTRK-like family, SLITRK1 0.028 -12.5 member 1 metastasis associated in colon MACC1 0.000 -12.3 cancer 1 cadherin-like and PC-esterase CPED1 0.029 -12.3 domain containing 1 gastrin-releasing peptide GRPR 0.006 -12.1 receptor LAMA1 0.001 -12.0 Se laminin, alpha 1 MAGEA12 0.006 -11.9 melanoma antigen family A, 12 TMC1 0.015 -11.6 transmembrane channel-like 1 JAG1 0.013 -11.3 B jagged 1 DIRAS family, GTP-binding DIRAS3 0.029 -11.1 RAS-like 3 RARB 0.003 -11.1 retinoic acid receptor, beta DSEL 0.004 -9.8 dermatan sulfate epimerase-like ITGB8 0.011 -9.8 integrin, beta 8 LHFP 0.002 -9.4 lipoma HMGIC fusion partner SORL1 0.003 -9.1 M E TES2 sortilin-related receptor, L galactose mutarotase (aldose 1- GALM 0.030 -8.6 M epimerase) RALY RNA binding protein- RALYL 0.000 -8.5 like solute carrier family 38, SLC38A4 0.001 -8.3 member 4 ASXL3 0.017 -8.1 additional sex combs like 3 SGCG 0.016 -7.6 sarcoglycan, gamma SULF2 0.021 -7.4 pB bcl3 sulfatase 2 GAGE2A 0.003 -7.2 pB bcl3 G antigen 2A TIMP3 0.010 -7.1 TIMP metallopeptidase inhibitor 3
Rho GTPase activating protein ARHGAP18 0.030 -7.1 Se 18 developmental pluripotency DPPA2 0.006 -7.1 associated 2 MYOF 0.003 -7.0 pB bcl3 myoferlin GAGE2E 0.001 -6.9 G antigen 2E BICC1 0.007 -6.9 bicaudal C homolog 1 ZNF804A 0.000 -6.8 CT1 bcl3 zinc finger protein 804A potassium channel, subfamily KCNK1 0.000 -6.8 K, member 1 GAGE2D 0.000 -6.7 G antigen 2D ATAD3C 0.000 -6.5 AAA domain containing 3C ANXA1 0.009 -6.3 M annexin A1 VGLL3 0.001 -6.3 vestigial like 3 RNF217 0.001 -6.2 ring finger protein 217 activated leukocyte cell ALCAM 0.003 -5.8 MSe TES2 adhesion molecule (CD166) HHIP 0.025 -5.8 hedgehog interacting protein FES 0.005 -5.7 M pB bcl3 feline sarcoma oncogene protein tyrosine phosphatase, PTPRD 0.001 -5.5 receptor type, D DACH2 0.014 -5.5 dachshund homolog 2 UDP-N-acetyl-alpha-D- galactosamine:polypeptide N- GALNT14 0.000 -5.4 M acetylgalactosaminyltransferase 14 S100 calcium binding protein S100A2 0.032 -5.4 BA A2 SLIT and NTRK-like family, SLITRK6 0.001 -5.4 Se member 6 EPHA8 0.043 -5.3 EPH receptor A8 Potassium inwardly-rectifying KCNJ8 0.037 -5.3 channel, subfamily J, member 8 teneurin transmembrane protein TENM1 0.009 -5.2 1 MAGEA1 0.016 -5.2 melanoma antigen family A aldehyde dehydrogenase 1 ALDH1A1 0.001 -5.2 family, member A1 PI15 0.001 -5.1 peptidase inhibitor 15 IQ motif containing with AAA IQCA1 0.029 -5.1 domain 1 fibroblast growth factor binding FGFBP3 0.000 -5.1 CT1 bcl3 protein 3 TFPI2 0.011 -5.0 A TES2 tissue factor pathway inhibitor 2 ABLIM1 0.001 -5.0 B CT1 p50 actin binding LIM protein 1 guanine nucleotide binding CT1 bcl3 protein (G protein), alpha GNAO1 0.041 -5.0 CT1 Stat3 activating activity polypeptide pB bcl3 O BCO2 0.019 -5.0 beta-carotene oxygenase 2 leucine rich repeat and coiled- LRRCC1 0.005 -5.0 S coil centrosomal protein 1 GAGE12D 0.033 -4.9 pB bcl3 G antigen 12D GAGE12J 0.044 -4.8 G antigen 12J tumor necrosis factor receptor TNFRSF19 0.004 -4.6 S Se CT1 bcl3 superfamily, member 19 ADAMTSL3 0.003 -4.5 ADAMTS-like 3 PROK2 0.015 -4.3 S prokineticin 2 regulator of G-protein signaling RGS5 0.000 -4.2 pB bcl3 5 GAGE1 0.001 -4.2 G antigen 1 NAV1 0.005 -4.2 S neuron navigator 1 LOC100144602 0.039 -4.1 GAS1 0.022 -4.1 B growth arrest-specific 1 thioredoxin-related TMX4 0.034 -4.1 M TES2 transmembrane protein 4 ZNF431 0.004 -4.1 zinc finger protein 431 V-set and transmembrane VSTM2L 0.046 -4.1 domain containing 2 like LOC101059948 0.040 -4.0 pB bcl3 family with sequence similarity FAM133A 0.034 -4.0 133, member A STK32B 0.016 -4.0 serine/threonine kinase 32B PLXDC2 0.023 -3.9 plexin domain containing 2 ArfGAP with RhoGAP domain, ARAP2 0.001 -3.9 S TES2 ankyrin repeat and PH domain 2 HOXB8 0.003 -3.8 homeobox B8 armadillo repeat containing, X- ARMCX4 0.026 -3.8 S linked 4 family with sequence similarity FAM110C 0.014 -3.8 110, member C GAP43 0.005 -3.7 growth associated protein 43 GAGE12H 0.013 -3.7 pB bcl3 G antigen 12H ciliary neurotrophic factor CNTFR 0.028 -3.6 S receptor TFPI 0.039 -3.5 Se tissue factor pathway inhibitor DLX6 0.022 -3.5 TES2 distal-less homeobox 6 CDH19 0.000 -3.4 cadherin 19 microfibrillar associated protein MFAP5 0.000 -3.4 5 GAGE12I 0.001 -3.4 G antigen 12I NID1 0.037 -3.4 M nidogen 1 TOM1L2 0.008 -3.4 target of myb1-like 2 met proto-oncogene (hepatocyte MET 0.028 -3.4 M growth factor receptor) DSG2 0.011 -3.4 B desmoglein 2 FXYD domain containing ion FXYD5 0.037 -3.3 transport regulator 5 family with sequence similarity FAM46A 0.027 -3.3 MB 46, member A DCP1B 0.002 -3.3 decapping mRNA 1B eukaryotic translation initiation EIF2D 0.010 -3.3 factor 2D eukaryotic translation initiation EIF4E3 0.000 -3.3 CT1 bcl3 factor 4E family member 3 SDPR 0.005 -3.3 S serum deprivation response chromosome 11 open reading C11orf63 0.027 -3.3 CT1 bcl3 frame 63 USP44 0.001 -3.3 CT1 bcl3 ubiquitin specific peptidase 44 glucan (1,4-alpha-), branching GBE1 0.000 -3.3 K CT1 bcl3 enzyme 1 leucine rich repeat containing LRRC4B 0.041 -3.3 TES2 4B FERMT1 0.043 -3.2 B fermitin family member 1 guanosine monophosphate GMPR 0.002 -3.2 M reductase SNX7 0.032 -3.2 sorting nexin 7 protein phosphatase 4, PPP4R1L 0.015 -3.2 S regulatory subunit 1-like SH3 domain containing ring SH3RF1 0.001 -3.1 TES2 CT1 bcl3 finger 1 TRIB2 0.026 -3.1 M tribbles homolog 2 egl-9 family hypoxia-inducible EGLN3 0.010 -3.1 factor 3 ADAM metallopeptidase with ADAMTS1 0.010 -3.1 SeA thrombospondin type 1 motif, 1 GAGE7 0.002 -3.1 G antigen 7 centrosomal protein 170kDa CEP170P1 0.002 -3.0 pseudogene 1 long intergenic non-protein LINC00568 0.001 -3.0 CT1 bcl3 coding RNA 568 HOXA5 0.007 -3.0 B A pB bcl3 CT1 bcl3 homeobox A5 EPDR1 0.044 -3.0 ependymin related 1 ANPEP 0.038 -3.0 SeS Alanyl aminopeptidase SPRY4 0.033 -2.9 sprouty homolog 4 neurotrophic tyrosine kinase, NTRK3 0.007 -2.9 S receptor, type 3 SEMA3A 0.000 -2.9 semaphorin 3A KIRREL 0.003 -2.9 kin of IRRE like APLN 0.004 -2.9 S apelin CRAT 0.035 -2.8 CT1 bcl3 carnitine O-acetyltransferase EPHA5 0.007 -2.8 EPH receptor A5 ATPase, Ca++ transporting, ATP2B4 0.019 -2.8 B TES2 plasma membrane 4 ZNF462 0.005 -2.7 zinc finger protein 462 C1QTNF9B- 0.043 -2.7 C1QTNF9B antisense RNA 1 AS1 B3GAT1 0.009 -2.7 beta-1,3-glucuronyltransferase 1 GAGE12C 0.048 -2.7 pB bcl3 G antigen 12C GAGE12E 0.048 -2.7 pB bcl3 G antigen 12E CD36 molecule CD36 0.013 -2.7 (thrombospondin receptor) phorbol-12-myristate-13- PMAIP1 0.015 -2.6 SeB acetate-induced protein 1 growth factor receptor-bound GRB10 0.045 -2.6 S protein 10 creatine kinase, mitochondrial CKMT1A 0.041 -2.6 1A OLFM1 0.009 -2.6 S B CT1 bcl3 olfactomedin 1 quinolinate QPRT 0.044 -2.6 phosphoribosyltransferase leucine rich repeat containing LRRC16A 0.003 -2.5 16A FBXO8 0.049 -2.5 F-box protein 8 guanine nucleotide binding protein (G protein), alpha GNAL 0.000 -2.5 bcl3 com activating activity polypeptide, olfactory type OTOL1 0.000 -2.5 otolin 1 solute carrier family 9, SLC9A2 0.009 -2.5 subfamily A LOC729732 0.038 -2.4 TTBK1 0.042 -2.4 tau tubulin kinase 1 lysophosphatidylcholine LPCAT2 0.025 -2.4 TES2 acyltransferase 2 HOXB6 0.045 -2.4 homeobox B6 SLIT2 0.001 -2.3 pB bcl3 slit homolog 2 EMP2 0.005 -2.2 S epithelial membrane protein 2 V-set and transmembrane VSTM4 0.045 -2.2 S pB bcl3 domain containing 4 microtubule-associated protein MAP2 0.000 -2.2 MSe CT1 bcl3 2 S100 calcium binding protein S100A10 0.001 -2.2 B A10 TWISTNB 0.047 -2.2 pB bcl3 CT1 bcl3 TWIST neighbor HOXD1 0.038 -2.2 homeobox D1 family with sequence similarity CT1 p50 195, member A pseudogene LOC286467 0.006 -2.2 CT1 Stat3 (LOC286467), non-coding RNA LOC644936 0.026 -2.1 guanine nucleotide binding GNAT3 0.032 -2.1 protein, alpha transducing 3 FZD1 0.005 -2.1 frizzled family receptor 1 bcl3 com TEX14 0.013 -2.1 testis expressed 14 CT1 bcl3 nuclear receptor interacting NRIP1 0.001 -2.1 protein 1 cytochrome b5 type B (outer mitochondrial membrane) CYB5B 0.001 -2.1 M TES2 (CYB5B), nuclear gene encoding mitochondrial protein serpin peptidase inhibitor, clade SERPINE2 0.004 -2.0 A BS E (nexin, plasminogen activator inhibitor type 1), member 2 SRY (sex determining region SOX5 0.000 -2.0 Y)-box 5 fibronectin type III and ankyrin FANK1 0.022 -2.0 repeat domains 1 CCNJL 0.044 -2.0 cyclin J-like MDFI 0.042 -2.0 MyoD family inhibitor WLS 0.003 -2.0 wntless homolog SH3-domain GRB2-like 1 SH3GL1P2 0.009 -1.9 pseudogene 2 Pancreatic and duodenal PDX1 0.017 -1.9 homeobox 1 SLIT and NTRK-like family, SLITRK5 0.005 -1.9 member 5 solute carrier family 25 (mitochondrial SLC25A21 0.017 -1.9 S TES2 oxodicarboxylate carrier), member 21 LACE1 0.017 -1.9 lactation elevated 1 Insulin-like growth factor- IGFBP4 0.012 -1.9 binding protein 4 neurofilament, heavy NEFH 0.010 -1.9 polypeptide protein kinase, AMP-activated, PRKAA2 0.001 -1.9 TES2 pB bcl3 alpha 2 catalytic subunit DDAH1 0.004 -1.9 B bcl3 com dimethylarginine CT1 bcl3 dimethylaminohydrolase v-ets avian erythroblastosis ETS2 0.004 -1.9 virus E26 oncogene homolog 2 KRT18 0.013 -1.8 keratin 18 membrane magnesium MMGT1 0.033 -1.8 transporter 1 IL1R1 0.006 -1.8 interleukin 1 receptor, type I DiGeorge syndrome critical DGCR6 0.019 -1.8 M region gene 6
CDH18 0.009 -1.8 cadherin 18, type 2 DMKN 0.001 -1.8 dermokine SNAI1 0.046 -1.8 S snail family zinc finger 1 KLHL26 0.034 -1.8 kelch-like family member 26 guanine nucleotide binding GNAI1 0.001 -1.8 S TES2 protein (G protein), alpha inhibiting activity polypeptide 1 glycine amidinotransferase (L- FAM198B 0.010 -1.8 arginine:glycine amidinotransferase) adaptor-related protein complex AP1S3 0.044 -1.8 M 1, sigma 3 VIM 0.003 -1.8 MK vimentin SYNPO2 0.030 -1.8 synaptopodin 2 ADCY1 0.016 -1.8 pB bcl3 adenylate cyclase 1 CA12 0.024 -1.8 carbonic anhydrase XII CA11 0.025 -1.8 CT1 bcl3 carbonic anhydrase XI SWI/SNF related, matrix associated, actin dependent SMARCA4 0.035 -1.7 B pB bcl3 CT1 bcl3 regulator of chromatin, subfamily a, member 4 glycine amidinotransferase (L- Se GATM 0.048 -1.7 M arginine:glycine Ma amidinotransferase) coiled-coil domain containing CCDC113 0.034 -1.7 CT1 bcl3 113 ZNF558 0.019 -1.6 zinc finger protein 558 solute carrier family 2 SLC2A3 0.046 -1.6 M TES2 (facilitated glucose transporter), member 3 potassium channel, subfamily KCNK5 0.031 -1.6 CT1 bcl3 K, member 5 DnaJ (Hsp40) homolog, DNAJC12 0.048 -1.6 TES2 subfamily C, member 12 CT1 bcl3 lymphocyte-specific protein 1 LOC654342 0.045 -1.6 pB bcl3 CT1 p50 pseudogene CT1 Stat3 SIRPA 0.006 -1.6 S signal-regulatory protein alpha tetratricopeptide repeat domain TTC26 0.002 -1.6 CT1 p50 26 XYLB 0.001 -1.6 S CT1 bcl3 xylulokinase homolog CCND2 0.022 -1.6 KBS cyclin D2 SLIT-ROBO Rho GTPase SRGAP1 0.002 -1.6 activating protein 1 bcl3 HIST1H2AM 0.013 -1.6 histone cluster 1, H2am com insulin-like growth factor IGFBP2 0.042 -1.6 B pB bcl3 CT1 bcl3 binding protein 2 microtubule associated MICAL2 0.032 -1.6 B CT1 p50 monoxygenase, calponin and LIM domain containing 2 CT1 bcl3 PH domain and leucine rich PHLPP1 0.040 -1.6 CT1 Stat3 repeat protein phosphatase 1 acyl-CoA synthetase long-chain ACSL3 0.012 -1.5 CT1 bcl3 family member 3 5-hydroxytryptamine HTR1D 0.024 -1.5 (serotonin) receptor 1D SYN1 0.007 -1.5 synapsin I coiled-coil-helix-coiled-coil- CHCHD10 0.038 -1.5 helix domain containing 10 NFASC 0.032 -1.5 bcl3 com neurofascin ATPase, Ca++ transporting, ATP2A2 0.014 -1.5 bcl3 com cardiac muscle, slow twitch 2 PEG10 0.013 -1.5 paternally expressed 10 cannabinoid receptor interacting CNRIP1 0.039 -1.5 S protein 1 KLF5 0.017 -1.5 MSe Kruppel-like factor 5 telomeric repeat binding factor TERF2 0.012 -1.5 M A pB bcl3 CT1 bcl3 2 WD repeat and FYVE domain WDFY1 0.016 -1.5 Se CT1 bcl3 containing 1 component of oligomeric golgi COG6 0.015 -1.5 TES2 complex 6 HIST1H2BD 0.013 -1.5 bcl3 com CT1 bcl3 histone cluster 1, H2bd coiled-coil domain containing CCDC50 0.003 -1.5 M TES2 50 catenin (cadherin-associated CTNNA3 0.003 -1.5 CT1 bcl3 protein), alpha 3 v-erb-b2 avian erythroblastic ERBB4 0.030 -1.5 CT1 bcl3 leukemia viral oncogene homolog 4 CT1 bcl3 ANKH 0.031 -1.5 TES2 CT1 bcl3 ankylosis, progressive homolog pB bcl3 PANK3 0.011 -1.5 CT1 bcl3 pantothenate kinase 3 solute carrier family 9, SLC9A6 0.017 -1.5 A CT1 bcl3 subfamily A (NHE6, cation proton antiporter 6), member 6 DEAH (Asp-Glu-Ala-His) box DHX32 0.018 -1.5 pB bcl3 polypeptide 32 trichoplein, keratin filament TCHP 0.039 -1.5 binding TOM1 0.044 -1.5 bcl3 com target of myb1 MCM3AP-AS1 0.035 -1.5 CT1 bcl3 MCM3AP antisense RNA 1 phospholipase B domain PLBD1 0.017 -1.5 containing 1 T-cell lymphoma invasion and TIAM1 0.040 -1.5 CT1 bcl3 metastasis 1 pterin-4 alpha-carbinolamine dehydratase/dimerization PCBD1 0.020 -1.5 CT1 bcl3 cofactor of hepatocyte nuclear factor 1 alpha
baculoviral IAP repeat BIRC3 0.005 182.8 A TES2 containing 3 B3GALT2 0.010 91.2 UDP-Gal:betaGlcNAc beta 1,3- galactosyltransferase, polypeptide 2 MIR31 host gene (non-protein MIR31HG 0.027 52.5 CT1 bcl3 coding) (MIR31HG), non- coding RNA SHH 0.027 50.8 sonic hedgehog ectonucleotide ENPP2 0.035 46.9 Se pyrophosphatase/phosphodieste rase 2 PRRX1 0.001 23.8 B paired related homeobox 1 CH25H 0.011 19.6 cholesterol 25-hydroxylase C19orf18 0.001 15.2 solute carrier family 7 (amino SLC7A7 0.015 14.8 M acid transporter light chain, y+L system), member 7 PLK2 0.004 14.6 S TES2 polo-like kinase 2 C8orf4 0.017 13.4 B prostaglandin-endoperoxide PTGS2 0.000 11.9 MSeB synthase 2 AREG 0.001 9.6 amphiregulin BMP and activin membrane- BAMBI 0.026 9.0 bound inhibitor epidermal growth factor EGFR 0.032 9.0 B receptor SERTAD4-AS1 0.038 8.6 SERTAD4 antisense RNA 1 regulator of G-protein signaling RGS2 0.007 8.5 B M CT1 bcl3 2 polycystic kidney disease 1 like PKD1L1 0.005 8.4 1 serpin peptidase inhibitor, clade SERPING1 0.004 8.2 G (C1 inhibitor), member 1 TRIM47 0.008 7.7 tripartite motif containing 47 nuclear factor of kappa light CT1 bcl3 NFKB2 0.003 6.6 EM TES2 polypeptide gene enhancer in B- CT1 bcl3 cells 2 (p49/p100) family with sequence similarity FAM229A 0.029 6.6 pB bcl3 CT1 bcl3 229, member A sterile alpha motif domain SAMD5 0.004 6.5 containing 5 papilin, proteoglycan-like PAPLN 0.000 6.3 TES2 pB bcl3 CT1 bcl3 sulfated glycoprotein CADM1 0.029 6.2 S cell adhesion molecule 1 CDC20B 0.034 6.2 CT1 bcl3 cell division cycle 20B G protein-coupled receptor, GPRC5A 0.044 6.2 family C, group 5, member A ACTA2 0.022 5.9 E actin, alpha 2 interferon induced with helicase IFIH1 0.000 5.7 C domain 1 FAT3 0.011 5.6 FAT atypical cadherin 3 FGF20 0.045 5.4 fibroblast growth factor 20 APOL6 0.002 5.1 M apolipoprotein L, 6 SSPN 0.044 4.7 sarcospan EPHB1 0.019 4.7 K TES2 EPH receptor B1 EPHX2 0.006 4.6 epoxide hydrolase 2 NFKBIA 0.050 4.6 MEBA Ma TES2 CT1 bcl3 IKBα SERTAD4 0.046 4.5 SERTA domain containing 4 NPW 0.007 4.3 neuropeptide W receptor tyrosine kinase-like ROR1 0.020 4.3 TES2 orphan receptor 1 RDH10 0.016 4.2 Se retinol dehydrogenase 10 NMBR 0.032 4.2 neuromedin B receptor bcl3 com ITGAV 0.002 4.0 EB Ma TES2 integrin, alpha V CT1 bcl3 CFB 0.027 3.9 complement factor B DEAD (Asp-Glu-Ala-Asp) box DDX60L 0.039 4.0 M polypeptide 60-like ANKRD33B 0.003 3.9 TES2 CT1 bcl3 ankyrin repeat domain 33B ZNF781 0.046 3.9 zinc finger protein 781 ZNF600 0.011 3.8 zinc finger protein 600 inositol polyphosphate-1- INPP1 0.023 3.8 EBMa phosphatase mixed lineage kinase domain- MLKL 0.023 3.7 M like GNMT 0.020 3.7 Glycine N-methyltransferase glucosaminyl (N-acetyl) GCNT4 0.025 3.4 transferase 4, core 2 ZNF667 0.017 3.4 S zinc finger protein 667 HMX2 0.026 3.4 CT1 bcl3 H6 family homeobox 2 TMEM249 0.002 3.4 pB bcl3 CT1 bcl3 transmembrane protein 249 JAM2 0.021 3.3 TES2 bcl3 com junctional adhesion molecule 2 TMC4 0.031 3.3 S pB bcl3 CT1 bcl3 transmembrane channel-like 4 LOC100128252 0.040 3.2 CRIP1 0.002 3.2 M pB bcl3 cysteine-rich protein 1 CT1 bcl3 zinc finger and SCAN domain ZSCAN18 0.002 3.2 B pB bcl3 pB bcl3 containing 18 RBP1 0.021 3.2 retinol binding protein 1 leucine-rich repeat LGI family, LGI2 0.025 3.2 member 2 cartilage intermediate layer CILP2 0.044 3.1 TES2 protein 2 v-rel avian reticuloendotheliosis RELB 0.016 3.0 B viral oncogene homolog B RAB42, member RAS RAB42 0.004 3.0 CT1 bcl3 oncogene family tumor necrosis factor receptor TNFRSF6B 0.024 3.0 bcl3 com superfamily, member 6b, decoy nuclear factor of kappa light NFKBIZ 0.005 3.0 M TES2 bcl3 com polypeptide gene enhancer in B- cells inhibitor, zeta ZNF471 0.017 2.9 zinc finger protein 471 CD27-AS1 0.007 2.9 CD27 antisense RNA 1 ZNF788 0.029 2.9 zinc finger family member 788 ZNF468 0.022 2.9 TES2 zinc finger protein 468 ZNF816 0.009 2.8 zinc finger protein 816 ZFP28 0.007 2.8 ZFP28 zinc finger protein lectin, galactoside-binding, LGALS3 0.001 2.8 CT1 bcl3 soluble, 3 EF-hand domain family, EFHD2 0.033 2.8 CT1 bcl3 member D2 ZNF611 0.002 2.7 zinc finger protein 611 olfactory receptor, family 7, OR7E14P 0.015 2.7 subfamily E, member 14 pseudogene cylindromatosis (turban tumor CYLD 0.002 2.7 TES2 syndrome) GLRX2 0.030 2.6 CT1 bcl3 glutaredoxin 2 (GLRX2), nuclear gene encoding mitochondrial protein ADM 0.007 2.6 B adrenomedullin KITLG 0.029 2.6 B KIT ligand membrane-associated ring MARCH2 0.008 2.6 pB bcl3 CT1 bcl3 finger (C3HC4) 2, E3 ubiquitin protein ligase carbohydrate (chondroitin 4) CHST13 0.042 2.6 TES2 sulfotransferase 13 COL9A2 0.030 2.6 collagen, type IX, alpha 2 apolipoprotein B mRNA editing APOBEC3B 0.003 2.5 S M enzyme, catalytic polypeptide- like 3B endoplasmic reticulum ERAP2 0.031 2.5 TES2 aminopeptidase 2 F-box and leucine-rich repeat FBXL7 0.029 2.5 CT1 bcl3 protein 7 HIV-1 Tat specific factor 1 HTATSF1P2 0.001 2.5 pB bcl3 pseudogene 2 translin-associated factor X TSNAXIP1 0.022 2.5 interacting protein 1 NPTX1 0.016 2.5 neuronal pentraxin I C3orf72 0.033 2.4 GLI1 0.040 2.4 CT1 bcl3 GLI family zinc finger 1 poly (ADP-ribose) polymerase PARP8 0.006 2.4 M family, member 8 ribosomal protein L23a RPL23AP64 0.026 2.4 bcl3 com pseudogene 64 endogenous retrovirus group ERVMER34-1 0.002 2.4 MER34, member 1 odd-skipped related transciption OSR1 0.034 2.4 S factor 1 N4BPL2 intronic transcript 2 N4BP2L2-IT2 0.024 2.4 (non-protein coding) ZNF28 0.009 2.3 zinc finger protein 28 tumor protein p53 regulated TP53AIP1 0.005 2.3 pB bcl3 apoptosis inducing protein 1 ZNF606 0.003 2.3 CT1 bcl3 zinc finger protein 606 killer cell lectin-like receptor KLRAP1 0.041 2.3 subfamily A pseudogene 1 ZNF329 0.027 2.2 bcl3 com zinc finger protein 329 matrix metallopeptidase 14 MMP14 0.018 2.2 CT1 bcl3 (membrane-inserted) 5-hydroxytryptamine HTR7P1 0.001 2.2 (serotonin) receptor 7 pseudogene 1 SLAIN1 0.012 2.2 TES2 CT1 bcl3 SLAIN motif family, member 1 COTL1 0.010 2.2 CT1 bcl3 coactosin-like 1 ZNF525 0.031 2.1 zinc finger protein 525 heat shock protein, alpha- HSPB6 0.039 2.1 TES2 CT1 bcl3 crystallin-related, B6 EMB 0.015 2.1 CT1 bcl3 embigin COL3A1 0.017 2.1 B collagen, type III, alpha 1 TEX40 0.028 2.1 testis expressed 40 DEAD (Asp-Glu-Ala-Asp) box DDX58 0.001 2.1 S polypeptide 58 ZNF222 0.003 2.1 zinc finger protein 222 DGAT2 0.022 2.1 M CT1 Stat3 diacylglycerol O- acyltransferase 2 VASH2 0.043 2.1 B TES2 CT1 p50 vasohibin 2 LOC90246 0.005 2.1 LOC642366 0.020 2.1 CT1 bcl3 proteasome (prosome, PSMB9 0.036 2.1 M macropain) subunit, beta type, 9 ZNF160 0.001 2.0 CT1 bcl3 zinc finger protein 160 LOC389641 0.008 2.0 3'-phosphoadenosine 5'- PAPSS2 0.012 2.0 phosphosulfate synthase 2 staufen, RNA binding protein, STAU2 0.028 2.0 M pB bcl3 homolog 2 LARGE 0.014 2.0 CT1 bcl3 like-glycosyltransferase MAGI2-AS3 0.015 2.0 pB bcl3 MAGI2 antisense RNA 3 von Willebrand factor C and VWCE 0.031 2.0 CT1 bcl3 EGF domains methylthioadenosine MTAP 0.046 1.9 Se TES2 bcl3 com phosphorylase protein disulfide isomerase PDIA2 0.049 1.9 CT1 bcl3 family A, member 2 ZNF547 0.006 1.9 zinc finger protein 547 ZNF234 0.029 1.9 zinc finger protein 234 pB bcl3 CT1 bcl3 MGC16275 0.028 1.9 bcl3 com ZNF418 0.023 1.9 zinc finger protein 418 tumor necrosis factor receptor TNFRSF10A 0.013 1.9 S superfamily, member 10a RBM43 0.033 1.9 RNA binding motif protein 43 NMI 0.043 1.9 MA N-myc (and STAT) interactor LOC100128398 0.029 1.9 LOC401320 0.013 1.9 TSPAN13 0.023 1.9 A MB CT1 bcl3 tetraspanin 13 CT1 bcl3 SDC4 0.043 1.9 B TES2 syndecan 4 CT1 Stat3 MIR17HG 0.048 1.9 M CT1 bcl3 miR-17-92 cluster host gene RNF207 0.009 1.9 ring finger protein 207 high mobility group HMGN5 0.005 1.8 TES2 nucleosome binding domain 5 DUSP8 0.005 1.8 dual specificity phosphatase 8 solute carrier family 39 (zinc SLC39A8 0.006 1.8 B S TES2 transporter), member 8 latent transforming growth LTBP3 0.025 1.8 A CT1 bcl3 factor beta binding protein 3 C9orf85 0.018 1.8 ZNF671 0.027 1.8 zinc finger protein 671 progestin and adipoQ receptor PAQR6 0.001 1.8 bcl3 com family member VI C3orf58 0.016 1.8 CT1 bcl3 CSF1 0.042 1.8 E TES2 colony stimulating factor 1 CMT1A duplicated region CDRT1 0.003 1.8 transcript 1 zinc finger, DHHC-type ZDHHC11 0.025 1.8 containing 11 LIM and senescent cell antigen- LIMS2 0.001 1.8 pB bcl3 like domains 2 ribonuclease 4 or 2-5A- RNASEL 0.032 1.8 M TES2 dependent ribonuclease pleckstrin homology-like PHLDA1 0.008 1.8 bcl3 com domain, family A, member 1 family with sequence similarity FAM171B 0.017 1.8 TES2 CT1 bcl3 171, member B CT1 bcl3 GPR176 0.023 1.8 G protein-coupled receptor 176 pB bcl3 ATP-Binding Cassette, Sub- ABCB6 0.018 1.8 pB bcl3 CT1 bcl3 Family B (MDR/TAP), Member 6 ZNF117 0.000 1.8 TES2 zinc finger protein 117 LOC653160 0.005 1.8 pB bcl3 CT1 bcl3 zinc finger, GATA-like protein ZGLP1 0.033 1.8 1 RBM12B-AS1 0.001 1.8 CT1 bcl3 RBM12B antisense RNA 1 LOC641746 0.019 1.7 LOC100130691 0.023 1.7 aryl hydrocarbon receptor ARNTL2 0.002 1.7 MAB bcl3 com nuclear translocator-like 2 poly (ADP-ribose) polymerase PARP10 0.027 1.7 M CT1 bcl3 family, member 10 coiled-coil domain containing CCDC102A 0.029 1.7 102A LOC254100 0.009 1.7 TMEM200B 0.032 1.7 CT1 bcl3 transmembrane protein 200B ankyrin repeat and sterile alpha ANKS1B 0.050 1.7 CT1 bcl3 motif domain containing 1B WAS protein family homolog 5 WASH5P 0.004 1.7 pB bcl3 CT1 bcl3 pseudogene ZNF585B 0.043 1.7 zinc finger protein 585B ZNF517 0.049 1.7 TES2 bcl3 com zinc finger protein 517 cyclin-dependent kinase CDKN2C 0.038 1.7 CT1 bcl3 inhibitor 2C (p18, inhibits CDK4) interleukin-1 receptor- IRAK4 0.007 1.7 associated kinase 4 leucine-rich single-pass LSMEM1 0.007 1.7 membrane protein 1 IL4R 0.004 1.7 interleukin 4 receptor mitogen-activated protein MAP3K8 0.045 1.7 A CT1 bcl3 kinase kinase kinase 8 KIAA1804 0.045 1.7 S Mixed Lineage Kinase 4 DOK3 0.002 1.7 M docking protein 3 TCIRG1 0.041 1.7 M pB bcl3 T-cell, immune regulator 1 family with sequence similarity FAM111A 0.030 1.7 M 111, member A butyrophilin, subfamily 2, BTN2A2 0.035 1.7 member A2 bcl3 com GJA3 0.002 1.7 gap junction protein, alpha 3 CT1 bcl3 cytochrome b-245, alpha CYBA 0.039 1.7 E CT1 bcl3 polypeptide TMEM86B 0.005 1.6 S pB bcl3 transmembrane protein 86B ISL1 0.037 1.6 CT1 bcl3 ISL LIM homeobox 1 potassium channel KCTD11 0.005 1.6 M tetramerization domain containing 11 hect domain and RLD 2 HERC2P7 0.018 1.6 pseudogene 7 ZNF814 0.040 1.6 zinc finger protein 814 ZNF264 0.003 1.6 zinc finger protein 264 bcl3 com CKB 0.002 1.6 M S creatine kinase CT1 bcl3 ZNF224 0.012 1.6 zinc finger protein 224 long intergenic non-protein LINC00340 0.044 1.6 CT1 bcl3 coding RNA 340 transmembrane and TMTC2 0.049 1.6 tetratricopeptide repeat containing 2 DEAD/H (Asp-Glu-Ala- DDX11L2 0.042 1.6 CT1 bcl3 Asp/His) box helicase 11 like 2 AMH 0.025 1.6 bcl3 com anti-Mullerian hormone ZNF419 0.007 1.6 M zinc finger protein 419 ZNF550 0.007 1.6 zinc finger protein 550 COL27A1 0.036 1.6 Se pB bcl3 CT1 bcl3 collagen, type XXVII, alpha 1 TBC1 domain family, member TBC1D30 0.029 1.6 TES2 30 RGM domain family, member RGMB 0.018 1.6 CT1 bcl3 B tubulin tyrosine ligase-like TTLL3 0.040 1.6 bcl3 com family, member 3 long intergenic non-protein LINC00174 0.048 1.6 pB bcl3 coding RNA 174, TAPBP 0.015 1.6 E TES2 bcl3 com TAP binding protein (tapasin) SEC31B 0.043 1.6 CT1 bcl3 SEC31 homolog B ZNF256 0.043 1.6 zinc finger protein 256 FKBP7 0.036 1.6 CT1 bcl3 FK506 binding protein 7 cyclin-dependent kinase bcl3 com CDKN2B 0.031 1.6 TES2 inhibitor 2B (p15, inhibits CT1 bcl3 CDK4) ZNF548 0.022 1.6 S zinc finger protein 548 baculoviral IAP repeat BIRC2 0.011 1.6 M CT1 bcl3 pB bcl3 containing 2 solute carrier family 23 SLC23A2 0.006 1.6 CT1 bcl3 (nucleobase transporters), member 2 CT1 bcl3 PLXNB1 0.002 1.6 B plexin B1 pB bcl3 HSF4 0.025 1.6 CT1 bcl3 heat shock transcription factor 4 ZNF44 0.016 1.6 zinc finger protein 44 LMO7 0.027 1.6 Se LIM domain 7 G protein-coupled receptor GPR137C 0.015 1.6 S TES2 137C TRIM73 0.038 1.5 pB bcl3 CT1 bcl3 tripartite motif containing 73 TRIM74 0.038 1.5 pB bcl3 CT1 bcl3 tripartite motif containing 74 pleckstrin and Sec7 domain PSD3 0.020 1.5 SB TES2 CT1 bcl3 containing 3 MYCBPAP 0.031 1.5 MYCBP associated protein BMP8A 0.033 1.5 bone morphogenetic protein 8a ZNF577 0.032 1.5 pB bcl3 zinc finger protein 577 ZNF84 0.037 1.5 B CT1 bcl3 zinc finger protein 84 ArfGAP with GTPase domain, AGAP4 0.004 1.5 ankyrin repeat and PH domain 4 FOXD2 0.041 1.5 CT1 bcl3 forkhead box D2 transcription factor binding to TFE3 0.013 1.5 IGHM enhancer 3 WDR5B 0.041 1.5 WD repeat domain 5B dual specificity phosphatase 5 DUSP5P1 0.040 1.5 TES2 bcl3 com pseudogene 1 LOC642799 0.036 1.5 ZNF32-AS2 0.049 1.5 ZNF32 antisense RNA 2 ZFP112 0.020 1.5 Zinc Finger Protein 112 protein kinase, X-linked, PRKXP1 0.047 1.5 pseudogene 1 UPK3BL 0.003 1.5 pB bcl3 uroplakin 3B-like bcl3 com three prime repair exonuclease TREX1 0.048 1.5 CT1 Stat3 1 COL5A2 0.018 1.5 B collagen, type V, alpha 2 roundabout, axon guidance ROBO1 0.050 1.5 B pB bcl3 CT1 bcl3 receptor, homolog 1 bcl3 com ATG16 autophagy related 16- ATG16L2 0.011 1.5 bcl3 com like 2 ZNF470 0.006 1.5 pB bcl3 CT1 bcl3 zinc finger protein 470 zinc finger, DHHC-type ZDHHC23 0.018 1.5 TES2 pB bcl3 CT1 bcl3 containing 23 sulfotransferase family, SULT1A3 0.011 1.5 M cytosolic, 1A, phenol- preferring, member 3 sulfotransferase family, SULT1A4 0.011 1.5 M cytosolic, 1A, phenol- preferring, member 4 TMEM158 0.003 1.5 B TES2 bcl3 com transmembrane protein 158 ELM2 and Myb/SANT-like ELMSAN1 0.033 1.5 bcl3 com domain containing 1 TMEM184A 0.026 1.5 transmembrane protein 184A blood vessel epicardial BVES 0.000 1.5 substance bcl3 com KDM6B 0.014 1.5 M Lysine Demethylase 6B CT1 bcl3 ZNF91 0.006 1.5 A TES2 zinc finger protein 91 *array data M = LMP1+ GC B cells (37), K = LMP1+ B-cells (2), S = LMP1+ transgenes (33), E = C33a + CTAR1 (9),
Se = LMP1+ NPC vs LMP1- NPC (32), A = LMP1+ keratinocytes (27), Ma = EBV+ NPC (3), B = EBV+ NPC (1)
** HEK-293 + TES2 (CTAR2) (11)
*** ChIP by bcl3 with FDR>10 and strong peaks (peak apex averages 7.2 fold over IgG (range of 17-4) in CTAR1 and 3.0 fold over IgG (range 4-2) in pB) or ChIP by p50 and pStat3 in CTAR1 with FDR>10 and overlap ENCODE peak
(As reference, bcl3 peaks with FDR<10, CT1 peak apex averages 9.1 fold above background IgG (range of 20-4) and pB peak apex averages 3.0 fold above background IgG (range of 5-2))
† bcl3 binds to same site in pBabe and CTAR1 expressing cells