Supplementary Data
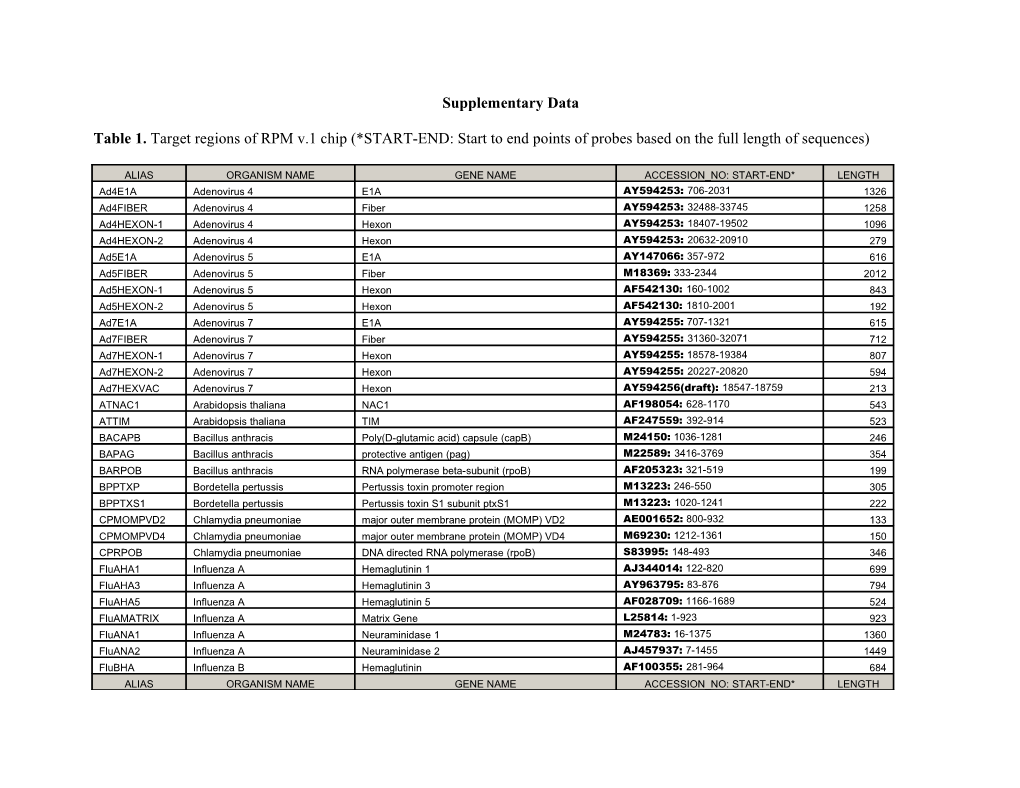
Table 1. Target regions of RPM v.1 chip (*START-END: Start to end points of probes based on the full length of sequences)
ALIAS ORGANISM NAME GENE NAME ACCESSION_NO: START-END* LENGTH Ad4E1A Adenovirus 4 E1A AY594253: 706-2031 1326 Ad4FIBER Adenovirus 4 Fiber AY594253: 32488-33745 1258 Ad4HEXON-1 Adenovirus 4 Hexon AY594253: 18407-19502 1096 Ad4HEXON-2 Adenovirus 4 Hexon AY594253: 20632-20910 279 Ad5E1A Adenovirus 5 E1A AY147066: 357-972 616 Ad5FIBER Adenovirus 5 Fiber M18369: 333-2344 2012 Ad5HEXON-1 Adenovirus 5 Hexon AF542130: 160-1002 843 Ad5HEXON-2 Adenovirus 5 Hexon AF542130: 1810-2001 192 Ad7E1A Adenovirus 7 E1A AY594255: 707-1321 615 Ad7FIBER Adenovirus 7 Fiber AY594255: 31360-32071 712 Ad7HEXON-1 Adenovirus 7 Hexon AY594255: 18578-19384 807 Ad7HEXON-2 Adenovirus 7 Hexon AY594255: 20227-20820 594 Ad7HEXVAC Adenovirus 7 Hexon AY594256(draft): 18547-18759 213 ATNAC1 Arabidopsis thaliana NAC1 AF198054: 628-1170 543 ATTIM Arabidopsis thaliana TIM AF247559: 392-914 523 BACAPB Bacillus anthracis Poly(D-glutamic acid) capsule (capB) M24150: 1036-1281 246 BAPAG Bacillus anthracis protective antigen (pag) M22589: 3416-3769 354 BARPOB Bacillus anthracis RNA polymerase beta-subunit (rpoB) AF205323: 321-519 199 BPPTXP Bordetella pertussis Pertussis toxin promoter region M13223: 246-550 305 BPPTXS1 Bordetella pertussis Pertussis toxin S1 subunit ptxS1 M13223: 1020-1241 222 CPMOMPVD2 Chlamydia pneumoniae major outer membrane protein (MOMP) VD2 AE001652: 800-932 133 CPMOMPVD4 Chlamydia pneumoniae major outer membrane protein (MOMP) VD4 M69230: 1212-1361 150 CPRPOB Chlamydia pneumoniae DNA directed RNA polymerase (rpoB) S83995: 148-493 346 FluAHA1 Influenza A Hemaglutinin 1 AJ344014: 122-820 699 FluAHA3 Influenza A Hemaglutinin 3 AY963795: 83-876 794 FluAHA5 Influenza A Hemaglutinin 5 AF028709: 1166-1689 524 FluAMATRIX Influenza A Matrix Gene L25814: 1-923 923 FluANA1 Influenza A Neuraminidase 1 M24783: 16-1375 1360 FluANA2 Influenza A Neuraminidase 2 AJ457937: 7-1455 1449 FluBHA Influenza B Hemaglutinin AF100355: 281-964 684 ALIAS ORGANISM NAME GENE NAME ACCESSION_NO: START-END* LENGTH FluBMATRIX Influenza B Matrix Gene AF100378: 13-374 362 FluBNA Influenza B Neuraminidase AY139081: 4-899 896 FTFOPA Francisella tularensis FopA AF097542: 767-877 111 FTLP Francisella tularensis 13-kDa lipoprotein M32059: 454-884 431 HCV229EMG Human coronavirus (229E) membrane glycoprotein AF304460: 24902-25499 598 HCVOC43MG Human coronavirus (OC43) membrane glycoprotein M93390: 211-568 358 HRV5NT Human rhinovirus 5’ noncoding region NC_001617: 168-579 412 LVGPC Lassa Virus GPC Gene M15076: 5-355 351 MPP1 Mycoplasma pneumoniae Cytadhesin P1 protei M18639: 4106-4474 369 NMCRGA Neisseria meningitidis regularoty protein, crgA AF190471: 499-752 254 NMCTRA Neisseria meningitidis capsular transport potein (ctrA) AE002098: 80991-81125 135 PIVIHN Parainfluenza Virus I HN U70948: 526-729 204 PIVIII5NCFP Parainfluenza Virus III 5' noncoding region Z11575: 4887-5116 230 PIVIIIHN Parainfluenza Virus III HN M18764: 705-917 213 RSVABL RSV L-polymerase AF254574: 2100-2478 379 RSVAN RSV A major nucleocapsid, M11486: 1099-1204 106 RSVBN RSV B major nucleocapsid, D00736: 1084-1211 128 SPNLYTA Streptococcus pneumoniae Autolysin, lytA AE007483: 9610-9734 125 SPNPLY Streptococcus pneumoniae pneumolysin, ply AE007483: 1562-1660 99 SPYERMB Streptococcus pyogenes erythromycin resistance methylase (ermB) X52632: 944-1191 248 SPYERMTR Streptococcus pyogenes erm(TR) AF002716: 379-554 176 SPYMEFAE Streptococcus pyogenes macrolide-efflux determinant (mefA, mefE) U70055: 370-739 370 SPYSPEB Streptococcus pyogenes pyrogenic exotoxin B (speB) L26155: 714-994 281 VMVCRMB Variola Major Virus cytokine response mo U88145: 262-552 291 VMVHA Variola Major Virus hemagglutinin (HA) X65516: 276-785 510 WNVCPRM West Nile virus C and prM AF196835: 233-664 432 WNVE West Nile virus E AF196835: 1160-1253 94 WNVNS1 West Nile virus NS1 AF196835: 3111-3263 153 YPCAF1 Yersinia pestis Caf1 X61996: 4629-5153 525 YPCVE Yersinia pestis cve2155 sequence AF350077: 34-298 265 ZEVL Ebola Virus L Gene AF086833: 13214-13656 443 29566 Table 2 Comparison of RPM v.1 results to culture results of clinical samples
Clinical sample RPM result Culture result NHRC 1 B. pertussis B. pertussis NHRC 2 B. pertussis B. pertussis NHRC 3 B. pertussis B. pertussis NHRC 4 B. pertussis B. pertussis NHRC 5 B. pertussis B. pertussis NHRC 6 B. pertussis B. pertussis NHRC 8 S. pneumoniae S. pneumoniae NHRC 10 S. pneumoniae S. pneumoniae NHRC 12 S. pneumoniae S. pneumoniae NHRC 14 S. pneumoniae S. pneumoniae NHRC 16 S. pneumoniae S. pneumoniae NHRC 18 S. pneumoniae S. pneumoniae NHRC 20 S. pneumoniae S. pneumoniae NHRC 22 S. pneumoniae S. pneumoniae 711863 HAdV-4 Adenovirus 719764 HAdV-4 Adenovirus “Permissive” Base Calling Algorithm Settings
– Filter Conditions
• No Signal threshold = 0.500 (default = 1.000000)
• Weak Signal Fold threshold = 20000.000 (default = 20.000000)
• Large SNR threshold = 20.000000 (default = 20.000000)
– Algorithm Parameters
• Strand Quality Threshold = 0.000 (default = 0.000000)
• Total Quality Threshold = 25.0000 (default = 75.000000)
• Maximum Fraction of Heterozygote Calls = 0.99000 (default = 0.900000)
• Model Type (0 = Heterozygote, 1 = Homozygote) = 0
• Perfect Call Quality Threshold = 0.500 (default = 2.000000)
– Final Reliability Rules
• Min Fraction of Calls in Neighboring Probes = 1.0000 (disables filter)
• Min Fraction of Calls of Samples = 1.0000 (disables filter)