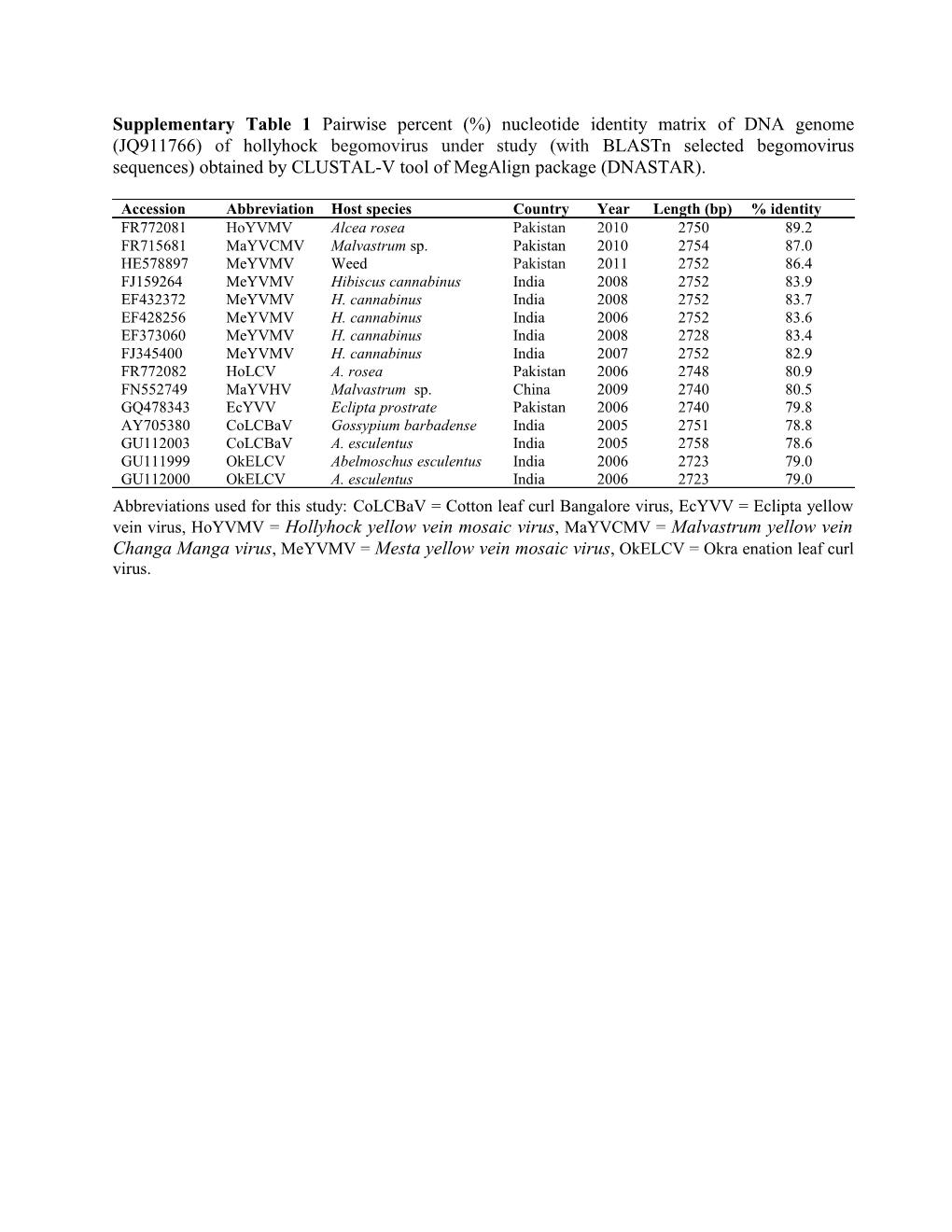
Supplementary Table 1 Pairwise percent (%) nucleotide identity matrix of DNA genome (JQ911766) of hollyhock begomovirus under study (with BLASTn selected begomovirus sequences) obtained by CLUSTAL-V tool of MegAlign package (DNASTAR).
Accession Abbreviation Host species Country Year Length (bp) % identity FR772081 HoYVMV Alcea rosea Pakistan 2010 2750 89.2 FR715681 MaYVCMV Malvastrum sp. Pakistan 2010 2754 87.0 HE578897 MeYVMV Weed Pakistan 2011 2752 86.4 FJ159264 MeYVMV Hibiscus cannabinus India 2008 2752 83.9 EF432372 MeYVMV H. cannabinus India 2008 2752 83.7 EF428256 MeYVMV H. cannabinus India 2006 2752 83.6 EF373060 MeYVMV H. cannabinus India 2008 2728 83.4 FJ345400 MeYVMV H. cannabinus India 2007 2752 82.9 FR772082 HoLCV A. rosea Pakistan 2006 2748 80.9 FN552749 MaYVHV Malvastrum sp. China 2009 2740 80.5 GQ478343 EcYVV Eclipta prostrate Pakistan 2006 2740 79.8 AY705380 CoLCBaV Gossypium barbadense India 2005 2751 78.8 GU112003 CoLCBaV A. esculentus India 2005 2758 78.6 GU111999 OkELCV Abelmoschus esculentus India 2006 2723 79.0 GU112000 OkELCV A. esculentus India 2006 2723 79.0 Abbreviations used for this study: CoLCBaV = Cotton leaf curl Bangalore virus, EcYVV = Eclipta yellow vein virus, HoYVMV = Hollyhock yellow vein mosaic virus, MaYVCMV = Malvastrum yellow vein Changa Manga virus, MeYVMV = Mesta yellow vein mosaic virus, OkELCV = Okra enation leaf curl virus.