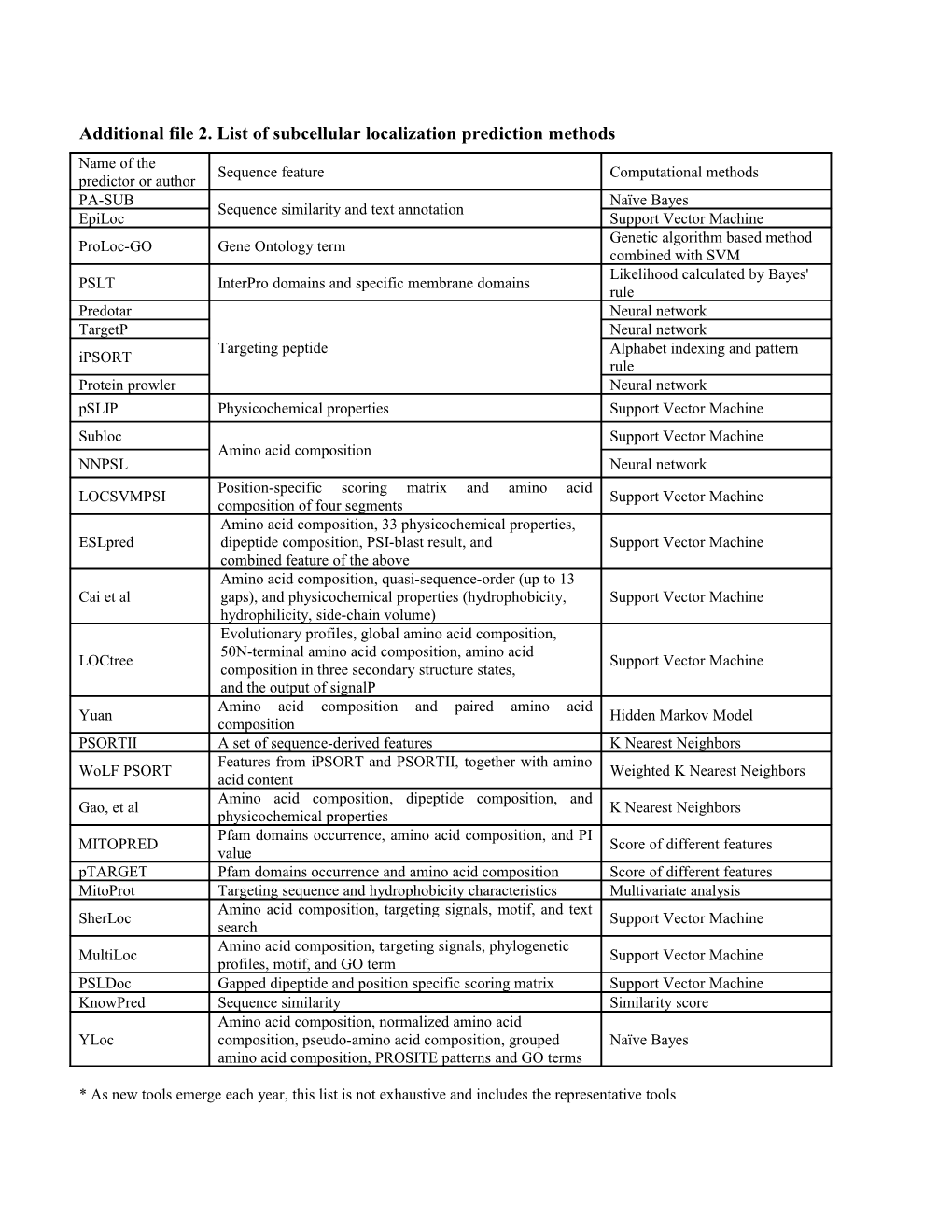
Additional file 2. List of subcellular localization prediction methods Name of the Sequence feature Computational methods predictor or author PA-SUB Naïve Bayes Sequence similarity and text annotation EpiLoc Support Vector Machine Genetic algorithm based method ProLoc-GO Gene Ontology term combined with SVM Likelihood calculated by Bayes' PSLT InterPro domains and specific membrane domains rule Predotar Neural network TargetP Neural network Targeting peptide Alphabet indexing and pattern iPSORT rule Protein prowler Neural network pSLIP Physicochemical properties Support Vector Machine Subloc Support Vector Machine Amino acid composition NNPSL Neural network Position-specific scoring matrix and amino acid LOCSVMPSI Support Vector Machine composition of four segments Amino acid composition, 33 physicochemical properties, ESLpred dipeptide composition, PSI-blast result, and Support Vector Machine combined feature of the above Amino acid composition, quasi-sequence-order (up to 13 Cai et al gaps), and physicochemical properties (hydrophobicity, Support Vector Machine hydrophilicity, side-chain volume) Evolutionary profiles, global amino acid composition, 50N-terminal amino acid composition, amino acid LOCtree Support Vector Machine composition in three secondary structure states, and the output of signalP Amino acid composition and paired amino acid Yuan Hidden Markov Model composition PSORTII A set of sequence-derived features K Nearest Neighbors Features from iPSORT and PSORTII, together with amino WoLF PSORT Weighted K Nearest Neighbors acid content Amino acid composition, dipeptide composition, and Gao, et al K Nearest Neighbors physicochemical properties Pfam domains occurrence, amino acid composition, and PI MITOPRED Score of different features value pTARGET Pfam domains occurrence and amino acid composition Score of different features MitoProt Targeting sequence and hydrophobicity characteristics Multivariate analysis Amino acid composition, targeting signals, motif, and text SherLoc Support Vector Machine search Amino acid composition, targeting signals, phylogenetic MultiLoc Support Vector Machine profiles, motif, and GO term PSLDoc Gapped dipeptide and position specific scoring matrix Support Vector Machine KnowPred Sequence similarity Similarity score Amino acid composition, normalized amino acid YLoc composition, pseudo-amino acid composition, grouped Naïve Bayes amino acid composition, PROSITE patterns and GO terms
* As new tools emerge each year, this list is not exhaustive and includes the representative tools