Table Interpretation and Extraction of Semantic Relationships to Synthesize Digital Documents
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Table Lens: Merging Graphical and Symbolic Representations in an Interactive Focus+ Context Visualization for Tabular Information
HumanFac(orsinComputingSystems CHI’94 0 “Celebra/i//ghrferdepende)~cc” The Table Lens: Merging Graphical and Symbolic Representations in an Interactive Focus+ Context Visualization for Tabular Information Ramana Rao and Stuart K. Card Xerox Palo Alto Research Center 3333 Coyote Hill Road Palo Alto, CA 94304 Gao,carcM@parc. xerox.com ABSTRACT (at cell size of 100 by 15 pixels, 82dpi). The Table Lens can We present a new visualization, called the Table Lens, for comfortably manage about 30 times as many cells and can visualizing and making sense of large tables. The visual- display up to 100 times as many cells in support of many ization uses a focus+ccmtext (fisheye) technique that works tasks. The scale advantage is obtained by using a so-called effectively on tabular information because it allows display “focus+context” or “fisheye” technique. These techniques of crucial label information and multiple distal focal areas. allow interaction with large information structures by dynam- In addition, a graphical mapping scheme for depicting table ically distorting the spatial layout of the structure according to contents has been developed for the most widespread kind the varying interest levels of its parts. The design of the Table of tables, the cases-by-variables table. The Table Lens fuses Lens technique has been guided by the particular properties symbolic and gaphical representations into a single coherent and uses of tables. view that can be fluidly adjusted by the user. This fusion and A second contribution of our work is the merging of graphical interactivity enables an extremely rich and natural style of representations directly into the process of table visualization direct manipulation exploratory data analysis. -

Information Extraction Based on Named Entity for Tourism Corpus
Information Extraction based on Named Entity for Tourism Corpus Chantana Chantrapornchai Aphisit Tunsakul Dept. of Computer Engineering Dept. of Computer Engineering Faculty of Engineering Faculty of Engineering Kasetsart University Kasetsart University Bangkok, Thailand Bangkok, Thailand [email protected] [email protected] Abstract— Tourism information is scattered around nowa- The ontology is extracted based on HTML web structure, days. To search for the information, it is usually time consuming and the corpus is based on WordNet. For these approaches, to browse through the results from search engine, select and the time consuming process is the annotation which is to view the details of each accommodation. In this paper, we present a methodology to extract particular information from annotate the type of name entity. In this paper, we target at full text returned from the search engine to facilitate the users. the tourism domain, and aim to extract particular information Then, the users can specifically look to the desired relevant helping for ontology data acquisition. information. The approach can be used for the same task in We present the framework for the given named entity ex- other domains. The main steps are 1) building training data traction. Starting from the web information scraping process, and 2) building recognition model. First, the tourism data is gathered and the vocabularies are built. The raw corpus is used the data are selected based on the HTML tag for corpus to train for creating vocabulary embedding. Also, it is used building. The data is used for model creation for automatic for creating annotated data. -

Army Acquisition Workforce Dependency on E-Mail for Formal
ARMY ACQUISITION WORKFORCE DEPENDENCY ON E-MAIL FOR FORMAL WORK COORDINATION: FINDINGS AND OPPORTUNITIES FOR WORKFORCE PERFORMANCE IMPROVEMENT THROUGH E-MAIL-BASED SOCIAL NETWORK ANALYSIS KENNETH A. LORENTZEN May 2013 PUBLISHED BY THE DEFENSE ACQUISITION UNIVERSITY PRESS PROJECT ADVISOR: BOB SKERTIC CAPITAL AND NORTHEAST REGION, DAU THE SENIOR SERVICE COLLEGE FELLOWSHIP PROGRAM ABERDEEN PROVING GROUND, MD PAGE LEFT BLANK INTENTIONALLY .ARMY ACQUISITION WORKFORCE DEPENDENCY ON E-MAIL FOR FORMAL WORK COORDINATION: FINDINGS AND OPPORTUNITIES FOR WORKFORCE PERFORMANCE IMPROVEMENT THROUGH E-MAIL-BASED SOCIAL NETWORK ANALYSIS KENNETH A. LORENTZEN May 2013 PUBLISHED BY THE DEFENSE ACQUISITION UNIVERSITY PRESS PROJECT ADVISOR: BOB SKERTIC CAPITAL AND NORTHEAST REGION, DAU THE SENIOR SERVICE COLLEGE FELLOWSHIP PROGRAM ABERDEEN PROVING GROUND, MD PAGE LEFT BLANK INTENTIONALLY ii Table of Contents Table of Contents ............................................................................................................................ ii List of Figures ................................................................................................................................ vi Abstract ......................................................................................................................................... vii Chapter 1—Introduction ................................................................................................................. 1 Background and Motivation ................................................................................................. -

Geotime As an Adjunct Analysis Tool for Social Media Threat Analysis and Investigations for the Boston Police Department Offeror: Uncharted Software Inc
GeoTime as an Adjunct Analysis Tool for Social Media Threat Analysis and Investigations for the Boston Police Department Offeror: Uncharted Software Inc. 2 Berkeley St, Suite 600 Toronto ON M5A 4J5 Canada Business Type: Canadian Small Business Jurisdiction: Federally incorporated in Canada Date of Incorporation: October 8, 2001 Federal Tax Identification Number: 98-0691013 ATTN: Jenny Prosser, Contract Manager, [email protected] Subject: Acquiring Technology and Services of Social Media Threats for the Boston Police Department Uncharted Software Inc. (formerly Oculus Info Inc.) respectfully submits the following response to the Technology and Services of Social Media Threats RFP. Uncharted accepts all conditions and requirements contained in the RFP. Uncharted designs, develops and deploys innovative visual analytics systems and products for analysis and decision-making in complex information environments. Please direct any questions about this response to our point of contact for this response, Adeel Khamisa at 416-203-3003 x250 or [email protected]. Sincerely, Adeel Khamisa Law Enforcement Industry Manager, GeoTime® Uncharted Software Inc. [email protected] 416-203-3003 x250 416-708-6677 Company Proprietary Notice: This proposal includes data that shall not be disclosed outside the Government and shall not be duplicated, used, or disclosed – in whole or in part – for any purpose other than to evaluate this proposal. If, however, a contract is awarded to this offeror as a result of – or in connection with – the submission of this data, the Government shall have the right to duplicate, use, or disclose the data to the extent provided in the resulting contract. GeoTime as an Adjunct Analysis Tool for Social Media Threat Analysis and Investigations 1. -

Inviwo — a Visualization System with Usage Abstraction Levels
IEEE TRANSACTIONS ON VISUALIZATION AND COMPUTER GRAPHICS, VOL X, NO. Y, MAY 2019 1 Inviwo — A Visualization System with Usage Abstraction Levels Daniel Jonsson,¨ Peter Steneteg, Erik Sunden,´ Rickard Englund, Sathish Kottravel, Martin Falk, Member, IEEE, Anders Ynnerman, Ingrid Hotz, and Timo Ropinski Member, IEEE, Abstract—The complexity of today’s visualization applications demands specific visualization systems tailored for the development of these applications. Frequently, such systems utilize levels of abstraction to improve the application development process, for instance by providing a data flow network editor. Unfortunately, these abstractions result in several issues, which need to be circumvented through an abstraction-centered system design. Often, a high level of abstraction hides low level details, which makes it difficult to directly access the underlying computing platform, which would be important to achieve an optimal performance. Therefore, we propose a layer structure developed for modern and sustainable visualization systems allowing developers to interact with all contained abstraction levels. We refer to this interaction capabilities as usage abstraction levels, since we target application developers with various levels of experience. We formulate the requirements for such a system, derive the desired architecture, and present how the concepts have been exemplary realized within the Inviwo visualization system. Furthermore, we address several specific challenges that arise during the realization of such a layered architecture, such as communication between different computing platforms, performance centered encapsulation, as well as layer-independent development by supporting cross layer documentation and debugging capabilities. Index Terms—Visualization systems, data visualization, visual analytics, data analysis, computer graphics, image processing. F 1 INTRODUCTION The field of visualization is maturing, and a shift can be employing different layers of abstraction. -

Extraction of Semantic Relations from Bioscience Text
Extraction of semantic relations from bioscience text by Barbara Rosario GRAD. (University of Trieste, Italy) 1995 A dissertation submitted in partial satisfaction of the requirements for the degree of Doctor of Philosophy in Information Management and Systems and the Designated Emphasis in Communication, Computation and Statistics in the GRADUATE DIVISION of the UNIVERSITY OF CALIFORNIA, BERKELEY Committee in charge: Professor Marti Hearst, Chair Professor John C. I. Chuang Professor Dan Klein Fall 2005 The dissertation of Barbara Rosario is approved: Professor Marti Hearst, Chair Date Professor John C. I. Chuang Date Professor Dan Klein Date University of California, Berkeley Fall 2005 Extraction of semantic relations from bioscience text Copyright c 2005 by Barbara Rosario Abstract Extraction of semantic relations from bioscience text by Barbara Rosario Doctor of Philosophy in Information Management and Systems and the Designated Emphasis in Communication, Computation and Statistics University of California, Berkeley Professor Marti Hearst, Chair A crucial area of Natural Language Processing is semantic analysis, the study of the meaning of linguistic utterances. This thesis proposes algorithms that ex- tract semantics from bioscience text using statistical machine learning techniques. In particular this thesis is concerned with the identification of concepts of interest (“en- tities”, “roles”) and the identification of the relationships that hold between them. This thesis describes three projects along these lines. First, I tackle the problem of classifying the semantic relations between nouns in noun compounds, to characterize, for example, the “treatment-for-disease” rela- tionship between the words of migraine treatment versus the “method-of-treatment” relationship between the words of sumatriptan treatment. -
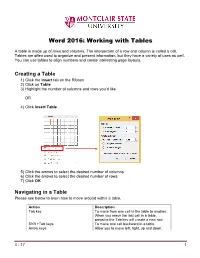
Word 2016: Working with Tables
Word 2016: Working with Tables A table is made up of rows and columns. The intersection of a row and column is called a cell. Tables are often used to organize and present information, but they have a variety of uses as well. You can use tables to align numbers and create interesting page layouts. Creating a Table 1) Click the Insert tab on the Ribbon 2) Click on Table 3) Highlight the number of columns and rows you’d like OR 4) Click Insert Table 5) Click the arrows to select the desired number of columns 6) Click the arrows to select the desired number of rows 7) Click OK Navigating in a Table Please see below to learn how to move around within a table. Action Description Tab key To move from one cell in the table to another. When you reach the last cell in a table, pressing the Tab key will create a new row. Shift +Tab keys To move one cell backward in a table. Arrow keys Allow you to move left, right, up and down. 4 - 17 1 Selecting All or Part of a Table There are times you want to select a single cell, an entire row or column, multiple rows or columns, or an entire table. Selecting an Individual Cell To select an individual cell, move the mouse to the left side of the cell until you see it turn into a black arrow that points up and to the right. Click in the cell at that point to select it. Selecting Rows and Columns To select a row in a table, move the cursor to the left of the row until it turns into a white arrow pointing up and to the right, as shown below. -

Table Understanding Through Representation Learning
TURL: Table Understanding through Representation Learning Xiang Deng∗ Huan Sun∗ Alyssa Lees The Ohio State University The Ohio State University Google Research Columbus, Ohio Columbus, Ohio New York, NY [email protected] [email protected] [email protected] You Wu Cong Yu Google Research Google Research New York, NY New York, NY [email protected] [email protected] ABSTRACT Relational tables on the Web store a vast amount of knowledge. Owing to the wealth of such tables, there has been tremendous progress on a variety of tasks in the area of table understanding. However, existing work generally relies on heavily-engineered task- specific features and model architectures. In this paper, we present TURL, a novel framework that introduces the pre-training/fine- tuning paradigm to relational Web tables. During pre-training, our framework learns deep contextualized representations on relational tables in an unsupervised manner. Its universal model design with pre-trained representations can be applied to a wide range of tasks with minimal task-specific fine-tuning. Specifically, we propose a structure-aware Transformer encoder to model the row-column structure of relational tables, and present a new Masked Entity Recovery (MER) objective for pre-training to capture the semantics and knowledge in large-scale unlabeled data. Figure 1: An example of a relational table from Wikipedia. We systematically evaluate TURL with a benchmark consisting of 6 different tasks for table understanding (e.g., relation extraction, a total of 14.1 billion tables in 2008. More recently, Bhagavatula et al. cell filling). We show that TURL generalizes well to all tasks and [4] extracted 1.6M high-quality relational tables from Wikipedia. -

VTK File Formats
File Formats for VTK Version 4.2 (Taken from The VTK User’s Guide Contact Kitware www.kitware.com to purchase) VTK File Formats The Visualization Toolkit provides a number of source and writer objects to read and write popular data file formats. The Visualization Toolkit also provides some of its own file formats. The main reason for creating yet another data file format is to offer a consistent data representation scheme for a variety of dataset types, and to provide a simple method to com- municate data between software. Whenever possible, we recommend that you use formats that are more widely used. But if this is not possible, the Visualization Toolkit formats described here can be used instead. Note that these formats may not be supported by many other tools. There are two different styles of file formats available in VTK. The simplest are the legacy, serial formats that are easy to read and write either by hand or programmatically. However, these formats are less flexible than the XML based file formats described later in this section. The XML formats support random access, parallel I/O, and portable data com- pression and are preferred to the serial VTK file formats whenever possible. Simple Legacy Formats The legacy VTK file formats consist of five basic parts. 1. The first part is the file version and identifier. This part contains the single line: # vtk DataFile Version x.x. This line must be exactly as shown with the exception of the version number x.x, which will vary with different releases of VTK. -

A Methodology for Engineering Domain Ontology Using Entity Relationship Model
(IJACSA) International Journal of Advanced Computer Science and Applications, Vol. 10, No. 8, 2019 A Methodology for Engineering Domain Ontology using Entity Relationship Model Muhammad Ahsan Raza1, M. Rahmah2, Sehrish Raza3, A. Noraziah4, Roslina Abd. Hamid5 Faculty of Computer Systems and Software Engineering, Universiti Malaysia Pahang, Kuantan, Malaysia1, 2, 4, 5 Institute of Computer Science and Information Technology, The Women University, Multan, Pakistan3 Abstract—Ontology engineering is an important aspect of The proposed procedure of ontology engineering (OE) is semantic web vision to attain the meaningful representation of novel in two aspects: (1) it is based on well know ER-schema data. Although various techniques exist for the creation of which is readily available for most of the database-based ontology, most of the methods involve the number of complex organizations or can be developed efficiently for any domain of phases, scenario-dependent ontology development, and poor interest with accuracy. (2) The method proposes instant and validation of ontology. This research work presents a lightweight cost-effective rules for ER to ontology translation while approach to build domain ontology using Entity Relationship maintaining all semantic checks. The ER-schema and (ER) model. Firstly, a detailed analysis of intended domain is translation model (viewing the two facets as simple and performed to develop the ER model. In the next phase, ER to portable) support the development of ontology for any ontology (EROnt) conversion rules are outlined, and finally the knowledge domain. Focusing on the discipline of information system prototype is developed to construct the ontology. The proposed approach investigates the domain of information technology where the learning material related to the technology curriculum for the successful interpretation of curriculum is highly unstructured, we have developed an OE concepts, attributes, relationships of concepts and constraints tool that captures semantics from the ER-schema and among the concepts of the ontology. -

000 Computer Science, Information, General Works
000 000 Computer science, information, general works SUMMARY 001–006 [Knowledge, the book, systems, computer science] 010 Bibliography 020 Library and information sciences 030 General encyclopedic works 050 General serial publications 060 General organizations and museology 070 Documentary media, educational media, news media; journalism; publishing 080 General collections 090 Manuscripts, rare books, other rare printed materials 001 Knowledge Description and critical appraisal of intellectual activity in general Including interdisciplinary works on consultants Class here discussion of ideas from many fields; interdisciplinary approach to knowledge Class epistemology in 121. Class a compilation of knowledge in a specific form with the form, e.g., encyclopedias 030 For consultants or use of consultants in a specific subject, see the subject, e.g., library consultants 023, engineering consultants 620, use of consultants in management 658.4 See Manual at 500 vs. 001 .01 Theory of knowledge Do not use for philosophy of knowledge, philosophical works on theory of knowledge; class in 121 .1 Intellectual life Nature and value For scholarship and learning, see 001.2 See also 900 for broad description of intellectual situation and condition 217 001 Dewey Decimal Classification 001 .2 Scholarship and learning Intellectual activity directed toward increase of knowledge Class methods of study and teaching in 371.3. Class a specific branch of scholarship and learning with the branch, e.g., scholarship in the humanities 001.3, in history 900 For research, -

Geotime Product Evaluation Lex Berman, CGA 12 Dec 09 Abstract
GeoTime Product Evaluation Lex Berman, CGA 12 Dec 09 Abstract: GeoTime is a stand-alone spatio-temporal browser with built-in queries for analyzing imported datasets containing events with defined spatial and temporal footprints. Events can be viewed as discrete points in a 3D space- time cube (with the the z-value representing the time axis), or can be viewed as entities (which are comprised of multiple location-events). The current version of GeoTime can directly load tables and flat files (including KML), but must be used together with ESRI ArcGIS to load from shapefiles and layers. Installation : The installation package for GeoTime includes an executable installer (for Windows only), a license file, and a third-party map library application, called MapMan. For GeoTime to run with ESRI files, both ArcGIS 9.2 or 9.3 must be installed, along with .NET, and Excel. Installation on ArcGIS 9.3 worked as expected during testing, but failed to completely integrate with ArcGIS 9.2 (due to an error with ESRI .NET assemblies). On the ArcGIS 9.3 install, the application worked for the administrator user profile only. It was not clear how to install the application for all user profiles. There are some options for installing the application with a license server, presumably to run as a keyed application over a limited subnet group of machines. However, since we are using the trial-period stand-alone client we don't know the level of difficulty in getting the license server (third-party software) to run. The MapMan package, included with GeoTime, takes considerably longer than GeoTime to install.