Differential Binding of Marvcf to Cadherins 3875
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

(COMT) Gene As a Candidate for Psychiatric Phenotypes
Molecular Psychiatry (2006) 11, 446–458 & 2006 Nature Publishing Group All rights reserved 1359-4184/06 $30.00 www.nature.com/mp FEATURE REVIEW The catechol-O-methyl transferase (COMT) gene as a candidate for psychiatric phenotypes: evidence and lessons N Craddock, MJ Owen and MC O’Donovan Department of Psychological Medicine, The Henry Wellcome Building for Biomedical Research in Wales, Cardiff University, School of Medicine, Heath Park, Cardiff, UK The enzyme catechol-O-methyl transferase (COMT), identified in the 1950s, is involved in catabolism of monoamines that are influenced by psychotropic medications, including neuroleptics and antidepressants. The COMT gene lies in a chromosomal region of interest for psychosis and bipolar spectrum disorder and a common polymorphism within the gene alters the activity of the enzyme. As a consequence, COMT has been one of the most studied genes for psychosis. On the basis of prior probabilities it would seem surprising if functional variation at COMT did not have some influence either on susceptibility to psychiatric phenotypes, modification of the course of illness or moderation of response to treatment. There is now robust evidence that variation at COMT influences frontal lobe function. However, despite considerable research effort, it has not proved straightforward to demonstrate and characterise a clear relationship between genetic variation at COMT and psychiatric phenotypes. It is of course, possible that COMT will turn out to be an unusually intractable case but it seems more likely that the experiences with this gene will provide a foretaste of the complexity of genotype–phenotype relationships that will be found for psychiatric traits. -

The N-Cadherin Interactome in Primary Cardiomyocytes As Defined Using Quantitative Proximity Proteomics Yang Li1,*, Chelsea D
© 2019. Published by The Company of Biologists Ltd | Journal of Cell Science (2019) 132, jcs221606. doi:10.1242/jcs.221606 TOOLS AND RESOURCES The N-cadherin interactome in primary cardiomyocytes as defined using quantitative proximity proteomics Yang Li1,*, Chelsea D. Merkel1,*, Xuemei Zeng2, Jonathon A. Heier1, Pamela S. Cantrell2, Mai Sun2, Donna B. Stolz1, Simon C. Watkins1, Nathan A. Yates1,2,3 and Adam V. Kwiatkowski1,‡ ABSTRACT requires multiple adhesion, cytoskeletal and signaling proteins, The junctional complexes that couple cardiomyocytes must transmit and mutations in these proteins can cause cardiomyopathies (Ehler, the mechanical forces of contraction while maintaining adhesive 2018). However, the molecular composition of ICD junctional homeostasis. The adherens junction (AJ) connects the actomyosin complexes remains poorly defined. – networks of neighboring cardiomyocytes and is required for proper The core of the AJ is the cadherin catenin complex (Halbleib and heart function. Yet little is known about the molecular composition of the Nelson, 2006; Ratheesh and Yap, 2012). Classical cadherins are cardiomyocyte AJ or how it is organized to function under mechanical single-pass transmembrane proteins with an extracellular domain that load. Here, we define the architecture, dynamics and proteome of mediates calcium-dependent homotypic interactions. The adhesive the cardiomyocyte AJ. Mouse neonatal cardiomyocytes assemble properties of classical cadherins are driven by the recruitment of stable AJs along intercellular contacts with organizational and cytosolic catenin proteins to the cadherin tail, with p120-catenin β structural hallmarks similar to mature contacts. We combine (CTNND1) binding to the juxta-membrane domain and -catenin β quantitative mass spectrometry with proximity labeling to identify the (CTNNB1) binding to the distal part of the tail. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -
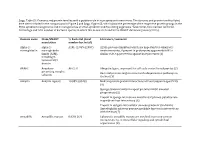
Supp. Table S2: Domains and Protein Families with a Putative Role in Host-Symbiont Interactions
Supp. Table S2: Domains and protein families with a putative role in host-symbiont interactions. The domains and protein families listed here were included in the comparisons in Figure 5 and Supp. Figure S5, which show the percentage of the respective protein groups in the Riftia symbiont metagenome and in metagenomes of other symbiotic and free-living organisms. % bacterial, total number bacterial: Percentage and total number of bacterial species in which this domain is found in the SMART database (January 2019). Domain name Pfam/SMART % bacterial (total Literature/comment annotation number bacterial) Alpha-2- alpha-2- A2M: 42.05% (2057) A2Ms: protease inhibitors which are important for eukaryotic macroglobulin macroglobulin innate immunity, if present in prokaryotes apparently fulfill a family (A2M), similar role, e.g. protection against host proteases (1) including N- terminal MG1 domain ANAPC Anaphase- APC2: 0 Ubiquitin ligase, important for cell cycle control in eukaryotes (2) promoting complex Bacterial proteins might interact with ubiquitination pathways in subunits the host (3) Ankyrin Ankyrin repeats 10.88% (8348) Mediate protein-protein interactions without sequence specificity (4) Sponge symbiont ankyrin-repeat proteins inhibit amoebal phagocytosis (5) Present in sponge microbiome metatranscriptomes, putative role in symbiont-host interactions (6) Present in obligate intracellular amoeba symbiont Candidatus Amoebophilus asiaticus genome, probable function in interactions with the host (7) Armadillo Armadillo repeats 0.83% (67) -

'Histology and Immunophenotype of Invasive Lobular Breast Cancer
Varga, Z; Mallon, E (2009). Histology and Immunophenotype of Invasive Lobular Breast Cancer. Daily Practice and Pitfalls. Breast Disease, 30:15-19. Postprint available at: http://www.zora.uzh.ch University of Zurich Posted at the Zurich Open Repository and Archive, University of Zurich. Zurich Open Repository and Archive http://www.zora.uzh.ch Originally published at: Breast Disease 2009, 30:15-19. Winterthurerstr. 190 CH-8057 Zurich http://www.zora.uzh.ch Year: 2009 Histology and Immunophenotype of Invasive Lobular Breast Cancer. Daily Practice and Pitfalls Varga, Z; Mallon, E Varga, Z; Mallon, E (2009). Histology and Immunophenotype of Invasive Lobular Breast Cancer. Daily Practice and Pitfalls. Breast Disease, 30:15-19. Postprint available at: http://www.zora.uzh.ch Posted at the Zurich Open Repository and Archive, University of Zurich. http://www.zora.uzh.ch Originally published at: Breast Disease 2009, 30:15-19. Histology and Immunophenotype of Invasive Lobular Breast Cancer. Daily Practice and Pitfalls Abstract Invasive lobular carcinomas (ILC) represent the most common subtype of invasive breast cancer and account for about 5-15% of all breast cancer cases. Invasive lobular carcinoma is often accompanied by in situ lesions, by lobular neoplasia (LN). Invasive lobular carcinomas display diverse histologic patterns varying from classical through solid to pleomorphic subtypes. When analyzing histological subtypes, the classical variant is reported to have a more favorable outcome. The majority of invasive lobular carcinomas are hormone receptor positive, overexpression and/or amplification of the Her2 gene is lower than in carcinomas of invasive ductal type. Loss of heterozygosity of the 16q chromosomal regions and the consequent lack of E-Cadherin expression are common findings in invasive lobular carcinomas. -

CDH1 Gene Cadherin 1
CDH1 gene cadherin 1 Normal Function The CDH1 gene provides instructions for making a protein called epithelial cadherin or E-cadherin. This protein is found within the membrane that surrounds epithelial cells, which are the cells that line the surfaces and cavities of the body, such as the inside of the eyelids and mouth. E-cadherin belongs to a family of proteins called cadherins whose function is to help neighboring cells stick to one another (cell adhesion) to form organized tissues. Another protein called p120-catenin, produced from the CTNND1 gene, helps keep E-cadherin in its proper place in the cell membrane, preventing it from being taken into the cell through a process called endocytosis and broken down prematurely. E-cadherin is one of the best-understood cadherin proteins. In addition to its role in cell adhesion, E-cadherin is involved in transmitting chemical signals within cells, controlling cell maturation and movement, and regulating the activity of certain genes. Interactions between the E-cadherin and p120-catenin proteins, in particular, are thought to be important for normal development of the head and face (craniofacial development), including the eyelids and teeth. E-cadherin also acts as a tumor suppressor protein, which means it prevents cells from growing and dividing too rapidly or in an uncontrolled way. Health Conditions Related to Genetic Changes Breast cancer Inherited mutations in the CDH1 gene increase a woman's risk of developing a form of breast cancer that begins in the milk-producing glands (lobular breast cancer). In many cases, this increased risk occurs as part of an inherited cancer disorder called hereditary diffuse gastric cancer (HDGC) (described below). -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Studies of Epigenetic Deregulation in Parathyroid Tumors and Small Intestinal Neuroendocrine Tumors
Digital Comprehensive Summaries of Uppsala Dissertations from the Faculty of Medicine 1377 Studies of epigenetic deregulation in parathyroid tumors and small intestinal neuroendocrine tumors ELHAM BARAZEGHI ACTA UNIVERSITATIS UPSALIENSIS ISSN 1651-6206 ISBN 978-91-513-0097-9 UPPSALA urn:nbn:se:uu:diva-330810 2017 Dissertation presented at Uppsala University to be publicly examined in Fåhraeussalen, Rudbecklaboratoriet Hus 5, Dag Hammarskjölds väg 20, Uppsala, Friday, 24 November 2017 at 13:15 for the degree of Doctor of Philosophy (Faculty of Medicine). The examination will be conducted in English. Faculty examiner: Associate Professor Christofer Juhlin (Department of Oncology-Pathology, Karolinska Institute). Abstract Barazeghi, E. 2017. Studies of epigenetic deregulation in parathyroid tumors and small intestinal neuroendocrine tumors. Digital Comprehensive Summaries of Uppsala Dissertations from the Faculty of Medicine 1377. 53 pp. Uppsala: Acta Universitatis Upsaliensis. ISBN 978-91-513-0097-9. Deregulation of the epigenome is associated with the initiation and progression of various types of human cancers. Here we investigated the level of 5-hydroxymethylcytosine (5hmC), expression and function of TET1 and TET2, and DNA methylation in parathyroid tumors and small intestinal neuroendocrine tumors (SI-NETs). In Paper I, an undetectable/very low level of 5hmC in parathyroid carcinomas (PCs) compared to parathyroid adenomas with positive staining, suggested that 5hmC may represent a novel biomarker for parathyroid malignancy. Immunohistochemistry revealed that increased tumor weight in adenomas was associated with a more aberrant staining pattern of 5hmC and TET1. A growth regulatory role of TET1 was demonstrated in parathyroid tumor cells. Paper II revealed that the expression of TET2 was also deregulated in PCs, and promoter hypermethylation was detected in PCs when compared to normal parathyroid tissues. -

Supplementary Materials
Supplementary materials Supplementary Table S1: MGNC compound library Ingredien Molecule Caco- Mol ID MW AlogP OB (%) BBB DL FASA- HL t Name Name 2 shengdi MOL012254 campesterol 400.8 7.63 37.58 1.34 0.98 0.7 0.21 20.2 shengdi MOL000519 coniferin 314.4 3.16 31.11 0.42 -0.2 0.3 0.27 74.6 beta- shengdi MOL000359 414.8 8.08 36.91 1.32 0.99 0.8 0.23 20.2 sitosterol pachymic shengdi MOL000289 528.9 6.54 33.63 0.1 -0.6 0.8 0 9.27 acid Poricoic acid shengdi MOL000291 484.7 5.64 30.52 -0.08 -0.9 0.8 0 8.67 B Chrysanthem shengdi MOL004492 585 8.24 38.72 0.51 -1 0.6 0.3 17.5 axanthin 20- shengdi MOL011455 Hexadecano 418.6 1.91 32.7 -0.24 -0.4 0.7 0.29 104 ylingenol huanglian MOL001454 berberine 336.4 3.45 36.86 1.24 0.57 0.8 0.19 6.57 huanglian MOL013352 Obacunone 454.6 2.68 43.29 0.01 -0.4 0.8 0.31 -13 huanglian MOL002894 berberrubine 322.4 3.2 35.74 1.07 0.17 0.7 0.24 6.46 huanglian MOL002897 epiberberine 336.4 3.45 43.09 1.17 0.4 0.8 0.19 6.1 huanglian MOL002903 (R)-Canadine 339.4 3.4 55.37 1.04 0.57 0.8 0.2 6.41 huanglian MOL002904 Berlambine 351.4 2.49 36.68 0.97 0.17 0.8 0.28 7.33 Corchorosid huanglian MOL002907 404.6 1.34 105 -0.91 -1.3 0.8 0.29 6.68 e A_qt Magnogrand huanglian MOL000622 266.4 1.18 63.71 0.02 -0.2 0.2 0.3 3.17 iolide huanglian MOL000762 Palmidin A 510.5 4.52 35.36 -0.38 -1.5 0.7 0.39 33.2 huanglian MOL000785 palmatine 352.4 3.65 64.6 1.33 0.37 0.7 0.13 2.25 huanglian MOL000098 quercetin 302.3 1.5 46.43 0.05 -0.8 0.3 0.38 14.4 huanglian MOL001458 coptisine 320.3 3.25 30.67 1.21 0.32 0.9 0.26 9.33 huanglian MOL002668 Worenine -

Somatic Mutational Landscapes of Adherens Junctions and Their
1 Somatic mutational landscapes of adherens junctions and their 2 functional consequences in cutaneous melanoma development 3 4 Praveen Kumar Korla,1 Chih-Chieh Chen,2 Daniel Esguerra Gracilla,1 Ming-Tsung Lai,3 Chih- 5 Mei Chen,4 Huan Yuan Chen,5 Tritium Hwang,1 Shih-Yin Chen,4,6,* Jim Jinn-Chyuan Sheu1,4, 6-9,* 6 1Institute of Biomedical Sciences, National Sun Yat-sen University, Kaohsiung 80242, Taiwan; 7 2Institute of Medical Science and Technology, National Sun Yat-sen University, Kaohsiung 80424, 8 Taiwan; 3Department of Pathology, Taichung Hospital, Ministry of Health and Welfare, Taichung 9 40343, Taiwan; 4Genetics Center, China Medical University Hospital, Taichung 40447, Taiwan; 10 5Institute of Biomedical Sciences, Academia Sinica, Taipei 11574, Taiwan; 6School of Chinese 11 Medicine, China Medical University, Taichung 40402, Taiwan; 7Department of Health and 12 Nutrition Biotechnology, Asia University, Taichung 41354, Taiwan; 8Department of 13 Biotechnology, Kaohsiung Medical University, Kaohsiung 80708, Taiwan; 9Institute of 14 Biopharmaceutical Sciences, National Sun Yat-sen University, Kaohsiung 80242, Taiwan 15 16 PKK, CCC and DEG contributed equally to this study. 17 *Correspondence to: Dr. Shih-Yin Chen ([email protected]) at Genetics Center, China 18 Medical University Hospital, Taichung, 40447, TAIWAN; or Dr. Jim Jinn-Chyuan Sheu 19 ([email protected]) at Institute of Biomedical Sciences, National Sun Yat-sen 20 University, Kaohsiung City 80424, TAIWAN. 21 22 Running title: mutational landscape of cadherins in melanoma 1 23 Abstract 24 Cell-cell interaction in skin homeostasis is tightly controlled by adherens junctions (AJs). 25 Alterations in such regulation lead to melanoma development. -

Armc5 Deletion Causes Developmental Defects and Compromises T-Cell Immune Responses
ARTICLE Received 26 Jan 2016 | Accepted 4 Nov 2016 | Published 7 Feb 2017 DOI: 10.1038/ncomms13834 OPEN Armc5 deletion causes developmental defects and compromises T-cell immune responses Yan Hu 1,*, Linjiang Lao1,*, Jianning Mao1, Wei Jin1, Hongyu Luo1, Tania Charpentier2, Shijie Qi1, Junzheng Peng1, Bing Hu3, Mieczyslaw Martin Marcinkiewicz4, Alain Lamarre2 & Jiangping Wu1,5 Armadillo repeat containing 5 (ARMC5) is a cytosolic protein with no enzymatic activities. Little is known about its function and mechanisms of action, except that gene mutations are associated with risks of primary macronodular adrenal gland hyperplasia. Here we map Armc5 expression by in situ hybridization, and generate Armc5 knockout mice, which are small in body size. Armc5 knockout mice have compromised T-cell proliferation and differentiation into Th1 and Th17 cells, increased T-cell apoptosis, reduced severity of experimental autoimmune encephalitis, and defective immune responses to lymphocytic choriomeningitis virus infection. These mice also develop adrenal gland hyperplasia in old age. Yeast 2-hybrid assays identify 16 ARMC5-binding partners. Together these data indicate that ARMC5 is crucial in fetal development, T-cell function and adrenal gland growth homeostasis, and that the functions of ARMC5 probably depend on interaction with multiple signalling pathways. 1 Centre de recherche (CR), Centre hospitalier de l’Universite´ de Montre´al (CHUM), 900 Rue Saint Denis, Montre´al, Que´bec, Canada H2X 0A9. 2 Institut national de la recherche scientifique—Institut Armand-Frappier (INRS-IAF), 531 Boul. des Prairies, Laval, Que´bec, Canada H7V 1B7. 3 Anatomic Pathology, AmeriPath Central Florida, 4225 Fowler Ave. Tampa, Orlando, Florida 33617, USA. -

Discovery and Systematic Characterization of Risk Variants and Genes For
medRxiv preprint doi: https://doi.org/10.1101/2021.05.24.21257377; this version posted June 2, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. It is made available under a CC-BY 4.0 International license . 1 Discovery and systematic characterization of risk variants and genes for 2 coronary artery disease in over a million participants 3 4 Krishna G Aragam1,2,3,4*, Tao Jiang5*, Anuj Goel6,7*, Stavroula Kanoni8*, Brooke N Wolford9*, 5 Elle M Weeks4, Minxian Wang3,4, George Hindy10, Wei Zhou4,11,12,9, Christopher Grace6,7, 6 Carolina Roselli3, Nicholas A Marston13, Frederick K Kamanu13, Ida Surakka14, Loreto Muñoz 7 Venegas15,16, Paul Sherliker17, Satoshi Koyama18, Kazuyoshi Ishigaki19, Bjørn O Åsvold20,21,22, 8 Michael R Brown23, Ben Brumpton20,21, Paul S de Vries23, Olga Giannakopoulou8, Panagiota 9 Giardoglou24, Daniel F Gudbjartsson25,26, Ulrich Güldener27, Syed M. Ijlal Haider15, Anna 10 Helgadottir25, Maysson Ibrahim28, Adnan Kastrati27,29, Thorsten Kessler27,29, Ling Li27, Lijiang 11 Ma30,31, Thomas Meitinger32,33,29, Sören Mucha15, Matthias Munz15, Federico Murgia28, Jonas B 12 Nielsen34,20, Markus M Nöthen35, Shichao Pang27, Tobias Reinberger15, Gudmar Thorleifsson25, 13 Moritz von Scheidt27,29, Jacob K Ulirsch4,11,36, EPIC-CVD Consortium, Biobank Japan, David O 14 Arnar25,37,38, Deepak S Atri39,3, Noël P Burtt4, Maria C Costanzo4, Jason Flannick40, Rajat M 15 Gupta39,3,4, Kaoru Ito18, Dong-Keun Jang4,