Introduction to R Software and Applications What Is the R Project
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Texworks: Lowering the Barrier to Entry
TEXworks: Lowering the barrier to entry Jonathan Kew 21 Ireton Court Thame OX9 3EB England [email protected] 1 Introduction The standard TEXworks workflow will also be PDF-centric, using pdfT X and X T X as typeset- One of the most successful TEX interfaces in recent E E E years has been Dick Koch's award-winning TeXShop ting engines and generating PDF documents as the on Mac OS X. I believe a large part of its success has default formatted output. Although it will still be been due to its relative simplicity, which has invited possible to configure a processing path based on new users to begin working with the system with- DVI, newcomers to the TEX world need not be con- out baffling them with options or cluttering their cerned with DVI at all, but can generally treat TEX screen with controls and buttons they don't under- as a system that goes directly from marked-up text stand. Experienced users may prefer environments files to ready-to-use PDF documents. T Xworks includes an integrated PDF viewer, such as iTEXMac, AUCTEX (or on other platforms, E based on the Poppler library, so there is no need WinEDT, Kile, TEXmaker, or many others), with more advanced editing features and project man- to switch to an external program such as Acrobat, agement, but the simplicity of the TeXShop model xpdf, etc., to view the typeset output. The inte- has much to recommend it for the new or occasional grated viewer also allows it to support source $ user. -

R Course for the Nsos in the Arab Countries Part I: Introduction
R Course for the NSOs in the Arab countries Part I: Introduction Valentin Todorov1 1United Nations Industrial Development Organization, Vienna 18-20 May 2015 Todorov (UNIDO) R Course for the NSOs in the Arab countriesPart I: Introduction18-20 May 2015 1 / 1 Outline Todorov (UNIDO) R Course for the NSOs in the Arab countriesPart I: Introduction18-20 May 2015 2 / 1 About R Outline Todorov (UNIDO) R Course for the NSOs in the Arab countriesPart I: Introduction18-20 May 2015 3 / 1 About R What is R • R is a language and environment for statistical computing and graphics • R is based on the S language originally developed by John Chambers and colleagues at AT&T Bell Labs in the late 1970s and early 1980s • R (sometimes called "GNU S\ ) is free open source software licensed under the GNU general public license (GPL 2) • R was created by Robert Gentleman and Ross Ihaka at the University of Auckland as a test bed for trying out some ideas in statistical computing • R is formally known as The R Project for Statistical Computing: http://www.r-project.org Todorov (UNIDO) R Course for the NSOs in the Arab countriesPart I: Introduction18-20 May 2015 4 / 1 About R The R project • The R Project is an international collaboration of researchers in statistical computing. • There are roughly 20 members of the "R Core Team\ who maintain and enhance R. • Releases of the R environment are made through the CRAN (comprehensive R archive network) twice per year. • The software is released under a "free software\ license, which makes it possible for anyone to download and use it. -

Travels in TEX Land: Choosing a TEX Environment for Windows
The PracTEX Journal TPJ 2005 No 02, 2005-04-15 Rev. 2005-04-17 Travels in TEX Land: Choosing a TEX Environment for Windows David Walden The author of this column wanders through world of TEX, as a non-expert, reporting what he observes and learns, which hopefully will be interesting to other non-expert users of TEX. 1 Introduction This column recounts my experiences looking at and thinking about different ways TEX is set up for users to go through the document-composition to type- setting cycle (input and edit, compile, and view or print). First, I’ll describe my own experience randomly trying various TEX environments. I suspect that some other users have had a similar introduction to TEX; and perhaps other users have just used the environment that was available at their workplace or school. Then I’ll consider some categories for thinking about options in TEX setups. Last, I’ll suggest some follow-on steps. Since I use Microsoft Windows as my computer operating system, this note focuses on environments that are available for Windows.1 2 My random path to choosing a TEX environment 2 I started using TEX in the late 1990s. 1But see my offer in Section 4. 2 While I started using TEX, I switched from TEX to using LATEX as soon as I discovered LATEX existed. Since both TEX and LATEX are operated in the same way, I’ll mostly refer to TEX in this note, since that is the more basic system. c 2005 David C. Walden I don’t quite remember my first setup for trying TEX. -

How to Write a Paper and Format It Using LATEX
How To Write a Paper and Format it Using LATEX Jennifer E. Hoffman1, 2, ∗ 1Department of Physics, Harvard University, Cambridge, Massachusetts 02138, USA 2School of Engineering & Applied Sciences, Harvard University, Cambridge, Massachusetts 02138, USA (Dated: July 30, 2020) The goal of this document is to demonstrate how to write a paper. We walk through the process of outlining, writing, formatting in LATEX, making figures, referencing, and checking style and content. Source files are available at: http://hoffman.physics.harvard.edu/example-paper/. I. GETTING STARTED 8. Rewrite your abstract, taking into account what you have learned from the process of writing the paper. As You should start writing your paper while you are you fine-tune your abstract, you may find it helpful to working on your experiment. Prof. George Whitesides refer to Nature's instructions for writing an abstract says: \A paper is not just an archival device for stor- [2] and for clear communication more generally [3]. ing a completed research program; it is also a structure for planning your research in progress. If you clearly As you contemplate the paper you have just writ- understand the purpose and form of a paper, it can be ten, put yourself in the shoes of the reviewers (includ- immensely useful to you in organizing and conducting ing your collaborators). You already work many, many your research. A good outline for the paper is also a hours/week, and you don't really want to spend more good plan for the research program. You should write time reading this paper. So you're going to be very happy and rewrite these plans/outlines throughout the course if the figures are pretty, the text flows logically, the ref- of the research. -

Miktex Manual Revision 2.0 (Miktex 2.0) December 2000
MiKTEX Manual Revision 2.0 (MiKTEX 2.0) December 2000 Christian Schenk <[email protected]> Copyright c 2000 Christian Schenk Permission is granted to make and distribute verbatim copies of this manual provided the copyright notice and this permission notice are preserved on all copies. Permission is granted to copy and distribute modified versions of this manual under the con- ditions for verbatim copying, provided that the entire resulting derived work is distributed under the terms of a permission notice identical to this one. Permission is granted to copy and distribute translations of this manual into another lan- guage, under the above conditions for modified versions, except that this permission notice may be stated in a translation approved by the Free Software Foundation. Chapter 1: What is MiKTEX? 1 1 What is MiKTEX? 1.1 MiKTEX Features MiKTEX is a TEX distribution for Windows (95/98/NT/2000). Its main features include: • Native Windows implementation with support for long file names. • On-the-fly generation of missing fonts. • TDS (TEX directory structure) compliant. • Open Source. • Advanced TEX compiler features: -TEX can insert source file information (aka source specials) into the DVI file. This feature improves Editor/Previewer interaction. -TEX is able to read compressed (gzipped) input files. - The input encoding can be changed via TCX tables. • Previewer features: - Supports graphics (PostScript, BMP, WMF, TPIC, . .) - Supports colored text (through color specials) - Supports PostScript fonts - Supports TrueType fonts - Understands HyperTEX(html:) specials - Understands source (src:) specials - Customizable magnifying glasses • MiKTEX is network friendly: - integrates into a heterogeneous TEX environment - supports UNC file names - supports multiple TEXMF directory trees - uses a file name database for efficient file access - Setup Wizard can be run unattended The MiKTEX distribution consists of the following components: • TEX: The traditional TEX compiler. -

An Introduction to ESS + Xemacs for Windows Users of R
An Introduction to ESS + XEmacs for Windows Users of R John Fox∗ McMaster University Revised: 23 January 2005 1WhyUseESS+XEmacs? Emacs is a powerful and widely used programmer’s editor. One of the principal attractions of Emacs is its programmability: Emacs can be adapted to provide customized support for program- ming languages, including such features as delimiter (e.g., parenthesis) matching, syntax highlight- ing, debugging, and version control. The ESS (“Emacs Speaks Statistics”) package provides such customization for several common statistical computing packages and programming environments, including the S family of languages (R and various versions of S-PLUS), SAS, and Stata, among others. The current document introduces the use of ESS with R running under Microsoft Windows. For some Unix/Linux users, Emacs is more a way of life than an editor: It is possible to do almost everything from within Emacs, including, of course, programming, but also writing documents in mark-up languages such as HTML and LATEX; reading and writing email; interacting with the Unix shell; web browsing; and so on. I expect that this kind of generalized use of Emacs will not be attractive to most Windows users, who will prefer to use familiar, and specialized, tools for most of these tasks. There are several versions of the Emacs editor, the two most common of which are GNU Emacs, a product of the Free Software Foundation, and XEmacs, an offshoot of GNU Emacs. Both of these implementations are free and are available for most computing platforms, including Windows. In my opinion, based only on limited experience, XEmacs offers certain advantages to Windows users, such as easier installation, and so I will explain the use of ESS under this version of the Emacs editor.1 As I am writing this, there are several Windows programming editors that have been customized for use with R. -

A Brief Introduction to Latex.Pdf
ME 601A, 2016-17, SEM II IIT Kanpur What are TeX and LaTeX? • LaTeX is a typesetting systems suitable for producing scientific and mathematical documents — LaTeX enables authors to typeset and print their work at the highest typographical quality. — LaTeX is pronounced “Lay-te ch”. — LaTeX uses TeX formatter as its typesetting engine. • TeX is a program written by Donald Kunth for typesetting text and mathematical formulas Why LaTeX ? • Easy to use, especially for typing mathematical formulas • Portability (Windows, Unix, Mac) • Stability and interchangeability Why LaTeX ? contd... • High quality Most journals /conferences have their LaTeX styles One will be forced to use it, since everyone else around is using it. Why LaTeX ? contd... Documentation and forums A universal acceptance among researchers Error finding and troubleshooting are not difficult ∞ J[x(⋅),u(⋅)] = ∫ F (x(t),u(t),t)dt t0 References for LaTeX — The not so short introduction to LaTeX2e http://tobi.oetiker.ch/lshort/lshort.pdf — Comprehensive TeX archive network http://www.ctan.org/ — Beginning LaTeX http://www.cs.cornell.edu/Info/Misc/LaTeX-Tutorial/LaTeX- Home.html — Google Leslie Lamport H. Kopka Process to Create a Document Using LaTeX TeX input file Your source LaTeX file.tex document Run LaTeX program DVI file Device independent file.dvi output Run Device Driver Unix Commands Output file > latex file.tex runs latex file.ps or file.pdf > xdiv file.dvi previewer > dvips file.dvi creates .ps > pdflatex file.tex creates .pdf directly How to Setup LaTeX for Windows • Download and install MikTeX LaTeX package http://www.miktex.org/ • Install Ghostscript and Gsview http://pages.cs.wisc.edu/~ghost/ PS device driver … • Install Acrobat Reader • Install Editor — WinEdt For MAC Users http://www.winedt.com/ TeXShop — TexnicCenter iTexMac http://www.texniccenter.org/ Texmaker — Emacs, vi, etc. -

Latex in Twenty Four Hours
Plan Introduction Fonts Format Listing Tabbing Table Figure Equation Bibliography Article Thesis Slide A Short Presentation on Dilip Datta Department of Mechanical Engineering, Tezpur University, Assam, India E-mail: [email protected] / datta [email protected] URL: www.tezu.ernet.in/dmech/people/ddatta.htm Dilip Datta A Short Presentation on LATEX in 24 Hours (1/76) Plan Introduction Fonts Format Listing Tabbing Table Figure Equation Bibliography Article Thesis Slide Presentation plan • Introduction to LATEX Dilip Datta A Short Presentation on LATEX in 24 Hours (2/76) Plan Introduction Fonts Format Listing Tabbing Table Figure Equation Bibliography Article Thesis Slide Presentation plan • Introduction to LATEX • Fonts selection Dilip Datta A Short Presentation on LATEX in 24 Hours (2/76) Plan Introduction Fonts Format Listing Tabbing Table Figure Equation Bibliography Article Thesis Slide Presentation plan • Introduction to LATEX • Fonts selection • Texts formatting Dilip Datta A Short Presentation on LATEX in 24 Hours (2/76) Plan Introduction Fonts Format Listing Tabbing Table Figure Equation Bibliography Article Thesis Slide Presentation plan • Introduction to LATEX • Fonts selection • Texts formatting • Listing items Dilip Datta A Short Presentation on LATEX in 24 Hours (2/76) Plan Introduction Fonts Format Listing Tabbing Table Figure Equation Bibliography Article Thesis Slide Presentation plan • Introduction to LATEX • Fonts selection • Texts formatting • Listing items • Tabbing items Dilip Datta A Short Presentation on LATEX -

The Acrotex Education Bundle for Latex, Manual of Usage
AcroTEX Software Development Team The AcroTEX eDucation Bundle (AeB) D. P. Story Copyright © 1999-2013 [email protected] Prepared: December 12, 2013 http://www.acrotex.net Table of Contents Preface 9 1 Getting Started 10 1.1 Installing the Distribution ........................... 10 1.2 Installing aeb.js ................................ 11 1.3 Language Localizations ............................. 12 1.4 Sample Files and Articles ........................... 12 1.5 Package Requirements ............................. 12 1.6 LATEXing Your First File ............................. 13 • For pdftex and dvipdfm Users ...................... 13 • For Distiller Users .............................. 14 The Web Package 15 2 Introduction 15 2.1 Overview ..................................... 15 2.2 Package Requirements ............................. 15 3 Basic Package Options 16 3.1 Setting the Driver Option ........................... 16 3.2 The tight Option ................................ 17 3.3 The usesf Option ................................ 17 3.4 The draft Option ................................ 17 3.5 The nobullets Option ............................. 17 3.6 The unicode Option .............................. 17 3.7 The useui Option ................................ 17 3.8 The xhyperref Option ............................. 18 4 Setting Screen Size 18 4.1 Custom Design .................................. 18 4.2 Screen Design Options ............................. 19 4.3 \setScreensizeFromGraphic ........................ 19 4.4 Using \addtoWebHeight -

The Treasure Chest for Compatibility with Texpower and Seminar
TUGboat, Volume 22 (2001), No. 1/2 67 the concept of pdfslide, but completely rewritten The Treasure Chest for compatibility with texpower and seminar. ifsym: in fonts Fonts with symbols for alpinistic, electronic, mete- orological, geometric, etc., usage. A LATEX2ε pack- age simplifies usage. Packages posted to CTAN jas99_m.bst: in biblio/bibtex/contrib “What’s in a name?” I did not realize that Jan Update of jas99.bst,modifiedforbetterconfor- Tschichold’s typographic standards lived on in the mity to the American Meteorological Society. koma-script package often mentioned on usenet (in LaTeX WIDE: in nonfree/systems/win32/LaTeX_WIDE comp.text.tex) until I happened upon the listing A demonstration version of an integrated editor for it in a previous edition of “The Treasure Chest”. and shell for TEX— free for noncommercial use, but without registration, customization is disabled. This column is an attempt to give TEX users an on- : LAT X2ε macro package of simple, “little helpers” going glimpse of the trove which is CTAN. lhelp E converted into dtx format. Includes common units This is a chronological list of packages posted with preceding thinspaces, framed boxes, start new to CTAN between June and December 2000 with odd or even pages, draft markers, notes, condi- descriptive text pulled from the announcement and tional includes (including EPS files), and versions edited for brevity — however, all errors are mine. of enumerate and itemize which allow spacing to Packages are in alphabetic order and are listed only be changed. in the last month they were updated. Individual files makecmds Provides commands to make commands, envi- / partial uploads are listed under their own name if ronments, counters and lengths. -
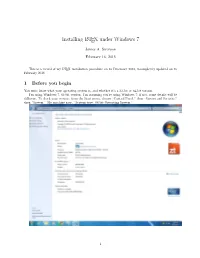
Installing LATEX Under Windows 7
Installing LATEX under Windows 7 James A. Swenson February 16, 2018 This is a record of my LATEX installation procedure on 18 December 2013, incompletely updated on 16 February 2018. 1 Before you begin You must know what your operating system is, and whether it's a 32-bit or 64-bit version. I'm using Windows 7, 64-bit version. I'm assuming you're using Windows 7; if not, some details will be different. To check your version: from the Start menu, choose \Control Panel," then \System and Security," then \System." My machine says, \System type: 64-bit Operating System." 1 2 The MiKTeX back end Go to http://miktex.org/download, or google \MiKTeX" and look for a download site. This site offers a \Recommended Download." I chose the \All Downloads" tab and clicked on \Basic MiKTeX 2.9.6615 64-bit Installer" rather than taking the recommended 32-bit version. Your version number will be slightly different. This file is large (∼210 MB); it may take a while to download. When the download finishes, run the file. I had to click \Yes" in a \User Account Control" box to give the program permission to run, then check a box accepting the copyright conditions and click \Next." I installed MiXTeX for all users (click \Next") and accepted the default installation directory (\Next"). I changed the default paper size to \Letter" and chose \Yes" for \Install missing packages on-the-fly." Clicking \Next" and \Start" got the process underway. This step took 2 or 3 minutes. When the green progress bars disappeared, I clicked \Next" and \Close." 3 Ghostscript Go to http://www.ghostscript.com/download/, or google \Ghostscript" and look for a download site. -

Research Techniques in Network and Information Technologies, February
Tools to support research M. Antonia Huertas Sánchez PID_00185350 CC-BY-SA • PID_00185350 Tools to support research The texts and images contained in this publication are subject -except where indicated to the contrary- to an Attribution- ShareAlike license (BY-SA) v.3.0 Spain by Creative Commons. This work can be modified, reproduced, distributed and publicly disseminated as long as the author and the source are quoted (FUOC. Fundació per a la Universitat Oberta de Catalunya), and as long as the derived work is subject to the same license as the original material. The full terms of the license can be viewed at http:// creativecommons.org/licenses/by-sa/3.0/es/legalcode.ca CC-BY-SA • PID_00185350 Tools to support research Index Introduction............................................................................................... 5 Objectives..................................................................................................... 6 1. Management........................................................................................ 7 1.1. Databases search engine ............................................................. 7 1.2. Reference and bibliography management tools ......................... 18 1.3. Tools for the management of research projects .......................... 26 2. Data Analysis....................................................................................... 31 2.1. Tools for quantitative analysis and statistics software packages ......................................................................................