Corporate Medical Policy
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

AMP Molecular Genetic Pathology Recommended Review Books
ASSOCIATION FOR MOLECULAR PATHOLOGY Education. Innovation & Improved Patient Care. Advocacy. 6120 Executive Blvd. Suite 700, Rockville, MD 20852 Tel: 301-634-7939 | Fax: 301-634-7995 | [email protected] | www.amp.org AMP Molecular Genetic Pathology : RECOMMENDED REVIEW BOOKS Updated: March, 2021 The following books are suggested for those preparing for the Molecular Genetic Pathology board exam administered by the American Board of Medical Genetics and the American Board of Pathology. This list represents the recommendations of the MGP Review Course faculty and the AMP Training and Education Committee; endorsement by the ABMG or the ABP is not implied. GENERAL MOLECULAR PATHOLOGY Thompson & Thompson Genetics in Medicine, 8th edition Nussbaum RL et al., eds. Elsevier, 2015 Molecular Pathology: The Molecular Basis of Human Diseases 2nd edition Coleman WB and Tsongalis GJ, eds. Elsevier, 2018 Diagnostic Molecular Pathology: A Guide to Applied Molecular Testing Coleman WB and Tsongalis GJ, eds. Elsevier, 2016 Molecular Basis of Human Cancer, 2nd edition Coleman WB and Tsongalis GJ, eds. Elsevier, 2017 Molecular Diagnostics: Fundamentals, Methods, and Clinical Applications 3rd edition Buckingham L. ASCP, 2021 Genomic Medicine: A Practical Guide Tafe LJ and Arcila ME, eds. Springer, 2020 CYTOGENETICS Gardner and Sutherland’s Chromosome Abnormalities and Genetic Counseling, 5th edition McKinlay Gardner RJ, Amor DJ. Oxford University Press, 2018 The Principles of Clinical Cytogenetics, 3rd edition Gersen SL and Keagle MB, eds. Human Press, 2013 Human Chromosomes, 4th edition Miller OJ and Therman E, eds. Springer Press, 2000 Vance GH, Khan WA. Utility of Fluorescence In Situ Hybridization in Clinical and Research Applications. Advances in Molecular Pathology 2020; 3:65-75. -

Whole Exome and Whole Genome Sequencing – Oxford Clinical Policy
UnitedHealthcare® Oxford Clinical Policy Whole Exome and Whole Genome Sequencing Policy Number: LABORATORY 024.11 T2 Effective Date: October 1, 2021 Instructions for Use Table of Contents Page Related Policies Coverage Rationale ....................................................................... 1 Chromosome Microarray Testing (Non-Oncology Documentation Requirements ...................................................... 2 Conditions) Definitions ...................................................................................... 2 Molecular Oncology Testing for Cancer Diagnosis, Prior Authorization Requirements ................................................ 3 Prognosis, and Treatment Decisions Applicable Codes .......................................................................... 3 • Preimplantation Genetic Testing Description of Services ................................................................. 4 Clinical Evidence ........................................................................... 4 U.S. Food and Drug Administration ........................................... 22 References ................................................................................... 22 Policy History/Revision Information ........................................... 26 Instructions for Use ..................................................................... 27 Coverage Rationale Whole Exome Sequencing (WES) Whole Exome Sequencing (WES) is proven and Medically Necessary for the following: • Diagnosing or evaluating a genetic disorder -

Whole Exome Sequencing Faqs
WHOLE EXOME SEQUENCING (WES) Whole Exome Sequencing (WES) is generally ordered when a patient’s medical history and physical exam strongly suggest that there is an underlying genetic etiology. In some cases, the patient may have had an extensive evaluation consisting of multiple genetic tests, without identifying an etiology. In other cases, a physician may opt to order one of the Whole Exome Sequencing tests early in the patient’s evaluation in an effort to expedite a possible diagnosis and reduce costs incurred by multiple tests. Whole Exome Sequencing is a highly complex test that is newly developed for the identification of changes in a patient’s DNA that are causative or related to their medical concerns. In contrast to current sequencing tests that analyze one gene or small groups of related genes at a time, Whole Exome Sequencing analyzes the exons or coding regions of thousands of genes simultaneously using next-generation sequencing techniques. The exome refers to the portion of the human genome that contains functionally important sequences of DNA that direct the body to make proteins essential for the body to function properly. These regions of DNA are referred to as exons. There are approximately 180,000 exons in the human genome which represents about 3% of the genome. These 180,000 exons are arranged in about 22,000 genes. It is known that many of the errors that occur in DNA sequences that then lead to genetic disorders are located in the exons. Therefore, sequencing of the exome is thought to be an efficient method of analyzing a patient’s DNA to discover the genetic cause of diseases or disabilities. -
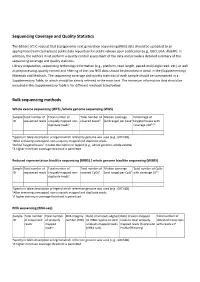
IJC Sequencing Coverage and Quality Statistics
Sequencing Coverage and Quality Statistics The Editors of IJC request that (epi)genomic next generation sequencing (NGS) data should be uploaded to an appropriate (restricted access) public data repository for public release upon publication (e.g., GEO, EGA, dbGAP). In addition, the authors must perform a quality control assessment of the data and provide a detailed summary of the sequencing coverage and quality statistics. Library preparation, sequencing technology information (e.g., platform, read length, paired‐end/single read, etc.) as well as preprocessing, quality control and filtering of the raw NGS data should be described in detail in the (Supplementary) Materials and Methods. The sequencing coverage and quality statistics of each sample should be summarized in a Supplementary Table, to which should be clearly referred in the main text. The minimum information that should be included in this Supplementary Table is for different methods listed below. Bulk sequencing methods Whole exome sequencing (WES) /whole genome sequencing (WGS) Sample Total number of Total number of Total number of Median coverage Percentage of ID sequenced reads uniquely mapped non‐ covered basesb (and range) per baseb targeted bases with duplicate readsa coverage ≥10b,c,d aSpecify in table description or legend which reference genome was used (e.g., GRCh38). bAfter removing unmapped, non‐uniquely mapped and duplicate reads. cDefine “targeted bases” in table description or legend (e.g., whole genome, whole exome). dA higher minimum coverage threshold is permitted. Reduced representation bisulfite sequencing (RRBS) / whole genome bisulfite sequencing (WGBS) Sample Total number of Total number of Total number of Median coverage Total number of CpGs ID sequenced reads uniquely mapped non‐ covered CpGsb (and range) per CpGb with coverage ≥5b,c duplicate readsa aSpecify in table description or legend which reference genome was used (e.g., GRCh38). -

Leave Policy for Residents and Fellows.Pdf
Leave Policy for Residents and Fellows The following policy outlines indications and training modifications to accommodate leave during accredited training for certification in all of the ABMGG specialties. Candidates for certification are required to successfully complete training in one of the following programs: ACGME-accredited Programs: 24 months (2 years) • Medical Genetics and Genomics Residency • Clinical Biochemical Genetics Postdoctoral Fellowship • Laboratory Genetics and Genomics Postdoctoral Fellowship Approved Combined Residency Programs: 48 months (4 years) • Medical Genetics and Genomics/Internal Medicine • Medical Genetics and Genomics/Pediatrics • Medical Genetics and Genomics/Maternal Fetal Medicine • Medical Genetics and Genomics/Reproductive Endocrinology and Infertility Subspecialty Fellowship Programs: 12 months (1 year) • Medical Biochemical Genetics • Molecular Genetic Pathology (cosponsored with American Board of Pathology) General Leave Policy: All candidates are allotted 4 weeks leave per year, which may be accrued or averaged through the duration of training for any indication. All candidates must take at least one week off per year as designated vacation time. Leave time cannot be forfeited to shorten training duration. Parental, Caregiver, and Medical Leave Policy: This policy refers to leave for the following indications: parental leave (the birth and care of a newborn including adopted and foster children for either partner), caregiver leave (care for an immediate (first degree) family member or partner with a serious health condition), or medical leave for significant personal medical needs. For Parental, Caregiver, and Medical leave, the ABMGG will allow up to 6 weeks leave during an academic year to occur once during the training program. For trainees who take indicated leave, they must still take at least one additional week for vacation during the same training year. -

The Economic Impact and Functional Applications of Human Genetics and Genomics
The Economic Impact and Functional Applications of Human Genetics and Genomics Commissioned by the American Society of Human Genetics Produced by TEConomy Partners, LLC. Report Authors: Simon Tripp and Martin Grueber May 2021 TEConomy Partners, LLC (TEConomy) endeavors at all times to produce work of the highest quality, consistent with our contract commitments. However, because of the research and/or experimental nature of this work, the client undertakes the sole responsibility for the consequence of any use or misuse of, or inability to use, any information or result obtained from TEConomy, and TEConomy, its partners, or employees have no legal liability for the accuracy, adequacy, or efficacy thereof. Acknowledgements ASHG and the project authors wish to thank the following organizations for their generous support of this study. Invitae Corporation, San Francisco, CA Regeneron Pharmaceuticals, Inc., Tarrytown, NY The project authors express their sincere appreciation to the following indi- viduals who provided their advice and input to this project. ASHG Government and Public Advocacy Committee Lynn B. Jorde, PhD ASHG Government and Public Advocacy Committee (GPAC) Chair, President (2011) Professor and Chair of Human Genetics George and Dolores Eccles Institute of Human Genetics University of Utah School of Medicine Katrina Goddard, PhD ASHG GPAC Incoming Chair, Board of Directors (2018-2020) Distinguished Investigator, Associate Director, Science Programs Kaiser Permanente Northwest Melinda Aldrich, PhD, MPH Associate Professor, Department of Medicine, Division of Genetic Medicine Vanderbilt University Medical Center Wendy Chung, MD, PhD Professor of Pediatrics in Medicine and Director, Clinical Cancer Genetics Columbia University Mira Irons, MD Chief Health and Science Officer American Medical Association Peng Jin, PhD Professor and Chair, Department of Human Genetics Emory University Allison McCague, PhD Science Policy Analyst, Policy and Program Analysis Branch National Human Genome Research Institute Rebecca Meyer-Schuman, MS Human Genetics Ph.D. -

Department of Molecular Biology and Immunology
Genetics Student Handbook 2020-2021 The information provided in this document serves to supplement the requirements of the Graduate School of Biomedical Sciences detailed in the UNTHSC Catalog with requirements specific to the Genetics discipline. Genetics Discipline (8/6/20) 1 Table of Contents Page Description of the Genetics Discipline ............................................ 3 Graduate Faculty and Their Research ............................................. 5 Requirements ................................................................................... 9 Required Courses ....................................................................... 9 Journal Club and Seminar Courses ............................................ 9 Elective Courses ......................................................................... 9 Sample Degree Plans ..................................................................... 11 Advancement to Candidacy ........................................................... 14 Genetics Discipline (8/6/20) 2 1. Description of the Genetics Discipline John V. Planz, Ph.D., Graduate Advisor Center for BioHealth, Rm 360 Phone: 817-735-2397 E-mail: [email protected] Program website: www.unthsc.edu/graduate-school-of-biomedical-sciences/genetics/ Graduate Faculty: Allen; Barber; Budowle; Cihlar; Coble; Ge; Phillips; Planz; Woerner Genetics is a broad interdisciplinary field that unites biochemistry, microbial and cellular biology, molecular processes, biotechnology, computational biology, biogeography and human disease -

Importance of Medical Genetics Research in Medicine
cs an eti d D n N e A G f R Journal of Genetics and DNA o e l s a e a n Sboui S, Tabbabi A, J Genet DNA Res 2018, 2:1 r r c u h o J Research Short Communication Open Access Importance of Medical Genetics Research in Medicine Sajida Sboui1 and Ahmed Tabbabi2* 1Faculty of Medicine of Monastir, Monastir University, Monastir, Tunisia 2Department of Hygiene and Environmental Protection, Ministry of Public Health, Tunis, Tunisia *Corresponding author: Ahmed Tabbabi, Department of Hygiene and Environmental Protection, Ministry of Public Health, Tunis, Tunisia, Tel: +216 71 577 115; +216 71 577 302; E-mail: [email protected] Received date: February 09, 2018, Accepted date: February 20, 2018, Published date: February 27, 2018 Copyright: © 2018 Sboui S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Keywords: Chromosome; Genes; Cleft lip; Transforming growth In medical genetics, the work diagnosis includes molecular analyzes factors (DNA, proteins and chromosomes) and clinical manisfestations of conditions, including malformations birth. If monogenic diseases are Short Communication rare, those caused by the gene/environment interaction are common and include many diseases. Prevention in medical genetics involves for About millions of families are affected by hereditary diseases around common pathologies the identification of subjects with high risk, with the world. Statistics analysis showed that about 5% of all pregnancies the intention of preventing the disease (e.g. -

Verification Activities: Sonia Salas
Verification Activities: New and Emerging Technologies Sonia Salas Verification Activities: New and Emerging Technologies • Verification: Methods, procedures, tests and other evaluations, in addition to monitoring, to determine whether a control measure or combination of control measures is or has been operating as intended and to establish the validity of the food safety plan (Source: FDA Industry Guidance: Control of Listeria monocytogenes in RTE Foods) • Verification activities include: validation, verification that monitoring is being conducted as required, verification of implementation and effectiveness, verification that appropriate decisions about corrective actions are being made and reanalysis (Source: PC Rule §117.155) Verification Activities: New and Emerging Technologies What new and emerging technologies can assist the produce sector in determining whether Listeria preventive controls are or have been operating as intended in a food safety plan? Verification Activities: New and Emerging Technologies Listeria controls (Source: FDA Industry Guidance: Control of Listeria monocytogenes in RTE Foods) • Controls on personnel • Design, construction and operation of a plant • Design, construction and maintenance of equipment • Sanitation • Storage practices and time/temperature controls • Transportation • Training • Controls on raw materials and ingredients • Process control based on formulating a RTE food • Listericial process control Verification Activities: New and Emerging Technologies – Whole Genome Sequencing (WGS) – Nanotechnology -

Next-Gen Sequencing Identifies Non-Coding Variation Disrupting
OPEN Molecular Psychiatry (2018) 23, 1375–1384 www.nature.com/mp ORIGINAL ARTICLE Next-gen sequencing identifies non-coding variation disrupting miRNA-binding sites in neurological disorders P Devanna1, XS Chen2,JHo1,2, D Gajewski1, SD Smith3, A Gialluisi2,4, C Francks2,5, SE Fisher2,5, DF Newbury6,7 and SC Vernes1,5 Understanding the genetic factors underlying neurodevelopmental and neuropsychiatric disorders is a major challenge given their prevalence and potential severity for quality of life. While large-scale genomic screens have made major advances in this area, for many disorders the genetic underpinnings are complex and poorly understood. To date the field has focused predominantly on protein coding variation, but given the importance of tightly controlled gene expression for normal brain development and disorder, variation that affects non-coding regulatory regions of the genome is likely to play an important role in these phenotypes. Herein we show the importance of 3 prime untranslated region (3'UTR) non-coding regulatory variants across neurodevelopmental and neuropsychiatric disorders. We devised a pipeline for identifying and functionally validating putatively pathogenic variants from next generation sequencing (NGS) data. We applied this pipeline to a cohort of children with severe specific language impairment (SLI) and identified a functional, SLI-associated variant affecting gene regulation in cells and post-mortem human brain. This variant and the affected gene (ARHGEF39) represent new putative risk factors for SLI. Furthermore, we identified 3′UTR regulatory variants across autism, schizophrenia and bipolar disorder NGS cohorts demonstrating their impact on neurodevelopmental and neuropsychiatric disorders. Our findings show the importance of investigating non-coding regulatory variants when determining risk factors contributing to neurodevelopmental and neuropsychiatric disorders. -

An Efficient and Scalable Analysis Framework for Variant Extraction and Refinement from Population-Scale DNA Sequence Data
Downloaded from genome.cshlp.org on October 4, 2021 - Published by Cold Spring Harbor Laboratory Press Method An efficient and scalable analysis framework for variant extraction and refinement from population-scale DNA sequence data Goo Jun,1,2 Mary Kate Wing,2 Gonçalo R. Abecasis,2 and Hyun Min Kang2 1Human Genetics Center, School of Public Health, The University of Texas Health Science Center at Houston, Houston, Texas 77030, USA; 2Center for Statistical Genetics and Department of Biostatistics, University of Michigan School of Public Health, Ann Arbor, Michigan 48109, USA The analysis of next-generation sequencing data is computationally and statistically challenging because of the massive vol- ume of data and imperfect data quality. We present GotCloud, a pipeline for efficiently detecting and genotyping high- quality variants from large-scale sequencing data. GotCloud automates sequence alignment, sample-level quality control, variant calling, filtering of likely artifacts using machine-learning techniques, and genotype refinement using haplotype in- formation. The pipeline can process thousands of samples in parallel and requires less computational resources than current alternatives. Experiments with whole-genome and exome-targeted sequence data generated by the 1000 Genomes Project show that the pipeline provides effective filtering against false positive variants and high power to detect true variants. Our pipeline has already contributed to variant detection and genotyping in several large-scale sequencing projects, including the 1000 Genomes Project and the NHLBI Exome Sequencing Project. We hope it will now prove useful to many medical sequencing studies. [Supplemental material is available for this article.] The cost of human genome sequencing has declined rapidly, pow- There is a pressing need for software pipelines that support ered by advances in massively parallel sequencing technologies. -

Clinical Whole Genome Sequencing
PRODUCT DATASHEET CLIA Certified and College of American Clinical Whole Pathologists (CAP) accredited Genome Built around our tried and tested PCR-Free Sequencing whole genome sequencing process OVERVIEW WHAT’S INCLUDED Clinical Human Whole Genome Sequencing provides validated, high quality sequence data generated through the same pro- • Sample receipt and nucleic acid extraction from blood cesses that have produced >100,000 research genomes - in- • QC & plating, sample preparation cluding genomes that have been included in large resource • Sample fidelity QC (96 SNP fingerprinting) datasets such as TOPMed and GnoMAD. Our in-house devel- oped, PCR-Free library construction process produces ex- • 2x150bp paired sequencing reads tremely even coverage of the entire genome, including exonic, • Coverage of ≥95% of bases ≥20x intronic, GC-rich, and regulatory regions. • Data delivery through a secure online portal Whole genome sequencing offers several advantages over tra- • Turnaround time ≤28 days from sample receipt ditional approaches of genomic characterization. Targeted pan- el and exome approaches require amplification of genomic DNA PERFORMANCE METRICS which induces sequence-context specific bias. Analyses of panels and exomes is limited to targeted regions and content % Bases at 20X Target: ≥95% must be updated if new regions are identified for study. Sever- Mean Coverage Target for Genome: ≥30X WGS Coverage al classes of human variation including copy number profiling and structural variation calling are more easily and accurately % Contamination: ≤2.5% (typically less than 0.2%) determined from whole genome sequencing. PF HQ Aligned Bases: ≥8x1010 bases Technical performance specifications for our clinical whole genome were determined through pre-validation analysis of Estimated Library Size: ≥7x109 molecules >20,000 research genomes as well as through input received SNV Sensitivity: 98.8% from our medical and population genetics community.