ANNUAL REPORT April 2012 to March 2013
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
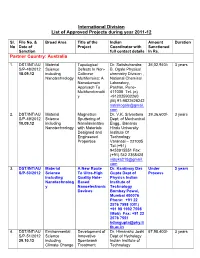
International Division List of Approved Projects During Year 2011-12
International Division List of Approved Projects during year 2011-12 Sl. File No. & Broad Area Title of the Indian Amount Duration No Date of Project Coordinator with Sanctioned Sanction full contact details In Rs. Partner Country: Australia 1. DST/INT/A U Material Topological Dr. Satishchandra 36,02,940/- 3 years S/P-48/2012 Science Defects In Non- B. Ogale Physical 18.09.12 including Collinear chemistry Division , Nanotechnology Multiferroics: A National Chemical Nanodomain Laboratory, Approach To Pashan, Pune- Multifunctionalit 411008 Tel. (o) y +912025902260 (M) 91-9822628242 satishogale@gmail. com 2. DST/INT/AU Material Magnetron Dr. V.K. Srivastava 39,36,600/- 3 years S/P-49/2012 Science Sputtering of Dept. of Mechanical 10.09.12 including Nanolaminates Engg., Banaras Nanotechnology with Materials Hindu University Designed and Institute Of Engineered Technology Properties Varanasi – 221005 Tel.(+91) 9453915551 Fax: (+91) 542 2368428 vijayks210@gmail. com 3. DST/INT/AU Material A New Route Dr. Kantimay Das Under 3 years S/P-50/2012 Science To Ultra-High Gupta Dept of Process including Quality Hole- Physics Indian Nanotechnolog Based Institute of y Nanoelectronic Technology Devices Bombay Powai, Mumbai 400076 Phone: +91 22 2576 7598 (Off.) +91 98 1992 7598 (Mob) Fax: +91 22 2576 7551 [email protected] tb.ac.in 4. DST/INT/AU Environmental Development of Dr. Himanshu Joshi 67,98,400/- 3 years S/P-51/2012 Science Innovative Dept of Hydrology 29.10.12 including Spentwash Indian Institute of Climate Change Treatment Technology Research Systems and Roorkee– 247667, Resource Uttarakhand Recovery himanshujoshi58@ Options for gmail.com; Distillery himanshu_joshi58 Application @yahoo.co.in; [email protected] n 5. -

Annual Report 2016 - 2017
Annual Report 2016 - 2017 CSIR-Centre for Cellular and Molecular Biology Hyderabad K Guruprasad 48 (Protein Sequence, Structure Analysis and Drug Design) K Thangaraj 50 (Evolutionary and Medical Genetics) Lakshmi Rao Kandukuri 54 (Chromosome Biology and Human Reproductive Genetics) Arvind Kumar 57 (Non-coding RNAs in diverse brain regions in stress response and depression) Lekha Dinesh Kumar 59 (Role of wnt signalling in EMT and development of colon cancer) Satish Kumar 62 (Functional Genomics using Transgenic and Knockout Mice and Molecular Approaches in Animal Breeding) Mukesh Lodha 65 (Mechanism of Epigenetic Inheritance in Plants) M M Idris 67 (Bio-mechanisms of Regeneration and Degeneration) M V Jagannadham 69 (Studies on outer membrane vesicles of bacteria) Rakesh K Mishra 72 (Genome Organization and Epigenetic Regulation) P Chandra Shekar 77 (Early embryonic development in mouse) Veena K Parnaik 79 (Nuclear organization and lamin biology) Anant B Patel 81 (13 C Nuclear Magnetic Resonance Investigations of Neurotransmitter Energetics in Neurological Disorders) R Nagaraj 85 Host-defense Antimicrobial Peptides; Activity and Developing Future Therapeutic Agents ii Palani Murugan Rangasamy 88 (The Regulation of Polyamine Homeostasis and their relevance in Health and Diseases of Eukaryotes) Ch Mohan Rao 90 (Molecular chaperones in health and diseases & Molecular diagnostics, therapeutics and drug delivery) Swasti Raychaudhuri 95 (Proteotoxicity in age-related diseases) Manjula Reddy 98 (Bacterial cell wall synthesis and its regulation) -

Centenary Celebration of Bose Institute
C E N T E N A R Y C E L E B R AT I O N CCEENNTTEENNAARRYY CCEELLEEBBRRAATTIIOONN OOFF BBOOSSEE IINNSSTTIITTUUTTEE 1 9 1 7 - 2 0 1 7 List of Posters (24.11.2017–28.11.2017) Poster Faculty Programme Title Poster Faculty Programme Title No No A-1 Amita Pal I Molecular characterization of VmMAPK1 and deciphering its role in restricting D-10 Tanya Das IV Is cancer a stem cell disease? MYMIV multiplication in tobacco Poulami Khan, Apoorva Bhattacharya, Shruti Banerjee, Swastika Paul, Abhishek Dutta, Anju Patel, Pankaj Kumar Singh, Shubho Chaudhuri and Amita Pal Dipanwita Dutta Chowdhury, Udit Basak, Apratim Dutta, Arijit Bhowmik, Devdutt A-2 Anupama Ghosh I Induction of apoptosis-like cell death and clearance of stress-induced intracellular Mazumdar, Aparajita Das, Sourio Chakraborty and Tanya Das protein aggregates: dual roles for Ustilago maydis metacaspase Mca1. E-1 Abhrajyoti Ghosh V Deciphering the code behind prokaryotic stress responses and ecophysiology A-1 Dibya Mukherjee, Sayandeep Gupta, Saran N, Rahul Datta, Anupama Ghosh Mousam Roy, Sayandeep Gupta, Chandrima Bhattacharyya, Shayantan Mukherji, A-3 Debabrata Basu I A multifaceted approach to unravel the signalling components of 'Black Spot' Abhrajyoti Ghosh A-2 disease resistance in oilseed mustard E-2 Srimonti Sarkar V The minimal ESCRT machinery of Giardia lamblia has altered inter-subunit Mrinmoy Mazumder, Amrita Mukherjee, Banani Mondal, Swagata Ghosh, Aishee De interactions within the ESCRT-II and ESCRT-III complexes A-3 and Debabrata Basu Nabanita Saha, Somnath Dutta and Srimonti Sarkar A-4 I Gaurab Gangyopadhyay Towards broadening the gene pool of few crop plants through molecular and E-3 Subrata Sau V Identification, purification and characterization of a cyclophilin from A-4 transgenic breeding Staphylococcus aureus Debabrata Dutta, Soumili Pal, Marufa Sultana, Vivek Arora and Gaurab Soham Seal, Debabrata Sinha, Subrata Sau Gangopadhyay E-4 Sujoy Kr. -

List of Life Members As on 20Th January 2021
LIST OF LIFE MEMBERS AS ON 20TH JANUARY 2021 10. Dr. SAURABH CHANDRA SAXENA(2154) ALIGARH S/O NAGESH CHANDRA SAXENA POST HARDNAGANJ 1. Dr. SAAD TAYYAB DIST ALIGARH 202 125 UP INTERDISCIPLINARY BIOTECHNOLOGY [email protected] UNIT, ALIGARH MUSLIM UNIVERSITY ALIGARH 202 002 11. Dr. SHAGUFTA MOIN (1261) [email protected] DEPT. OF BIOCHEMISTRY J. N. MEDICAL COLLEGE 2. Dr. HAMMAD AHMAD SHADAB G. G.(1454) ALIGARH MUSLIM UNIVERSITY 31 SECTOR OF GENETICS ALIGARH 202 002 DEPT. OF ZOOLOGY ALIGARH MUSLIM UNIVERSITY 12. SHAIK NISAR ALI (3769) ALIGARH 202 002 DEPT. OF BIOCHEMISTRY FACULTY OF LIFE SCIENCE 3. Dr. INDU SAXENA (1838) ALIGARH MUSLIM UNIVERSITY, ALIGARH 202 002 HIG 30, ADA COLONY [email protected] AVANTEKA PHASE I RAMGHAT ROAD, ALIGARH 202 001 13. DR. MAHAMMAD REHAN AJMAL KHAN (4157) 4/570, Z-5, NOOR MANZIL COMPOUND 4. Dr. (MRS) KHUSHTAR ANWAR SALMAN(3332) DIDHPUR, CIVIL LINES DEPT. OF BIOCHEMISTRY ALIGARH UP 202 002 JAWAHARLAL NEHRU MEDICAL COLLEGE [email protected] ALIGARH MUSLIM UNIVERSITY ALIGARH 202 002 14. DR. HINA YOUNUS (4281) [email protected] INTERDISCIPLINARY BIOTECHNOLOGY UNIT ALIGARH MUSLIM UNIVERSITY 5. Dr. MOHAMMAD TABISH (2226) ALIGARH U.P. 202 002 DEPT. OF BIOCHEMISTRY [email protected] FACULTY OF LIFE SCIENCES ALIGARH MUSLIM UNIVERSITY 15. DR. IMTIYAZ YOUSUF (4355) ALIGARH 202 002 DEPT OF CHEMISTRY, [email protected] ALIGARH MUSLIM UNIVERSITY, ALIGARH, UP 202002 6. Dr. MOHAMMAD AFZAL (1101) [email protected] DEPT. OF ZOOLOGY [email protected] ALIGARH MUSLIM UNIVERSITY ALIGARH 202 002 ALLAHABAD 7. Dr. RIAZ AHMAD(1754) SECTION OF GENETICS 16. -

PE PGRS33 Contributes to Mycobacterium Tuberculosis Entry in Macrophages Through Interaction with TLR2
RESEARCH ARTICLE PE_PGRS33 Contributes to Mycobacterium tuberculosis Entry in Macrophages through Interaction with TLR2 Ivana Palucci1, Serena Camassa1, Alessandro Cascioferro3, Michela Sali1, Saber Anoosheh3, Antonella Zumbo1, Mariachiara Minerva1, Raffaella Iantomasi1, Flavio De Maio1, Gabriele Di Sante2, Francesco Ria2, Maurizio Sanguinetti1, Giorgio Palù3, Michael J. Brennan4, Riccardo Manganelli3, Giovanni Delogu1* 1 Institute of Microbiology, Università Cattolica del Sacro Cuore, L.go A. Gemelli, 8–00168, Rome, Italy, 2 Institute of General Pathology, Università Cattolica del Sacro Cuore, L.go A. Gemelli, 8–00168, Rome, Italy, 3 Department of Molecular Medicine, University of Padua, Via A. Gabelli, 63–35121, Padua, Italy, 4 Aeras, Rockville (MD), United States of America * [email protected] Abstract PE_PGRS represent a large family of proteins typical of pathogenic mycobacteria whose members are characterized by an N-terminal PE domain followed by a large Gly-Ala repeat- OPEN ACCESS rich C-terminal domain. Despite the abundance of PE_PGRS-coding genes in the Myco- Citation: Palucci I, Camassa S, Cascioferro A, Sali bacterium tuberculosis (Mtb) genome their role and function in the biology and pathogene- M, Anoosheh S, Zumbo A, et al. (2016) PE_PGRS33 sis still remains elusive. In this study, we generated and characterized an Mtb H37Rv Contributes to Mycobacterium tuberculosis Entry in mutant (MtbΔ33) in which the structural gene of PE_PGRS33, a prototypical member of the Macrophages through Interaction with TLR2. PLoS ONE 11(3): e0150800. doi:10.1371/journal. protein family, was inactivated. We showed that this mutant entered macrophages with an pone.0150800 efficiency up to ten times lower than parental or complemented strains, while its efficiency in Editor: Joyoti Basu, Bose Institute, INDIA infecting pneumocytes remained unaffected. -

IL-32: an Emerging Player in the Immune Response Network Against Tuberculosis? Manikuntala Kundu, Joyoti Basu*
Perspectives IL-32: An Emerging Player in the Immune Response Network against Tuberculosis? Manikuntala Kundu, Joyoti Basu* nterleukin-32 (IL-32), which was Stimulation of IL-32 by Strengths and Weaknesses previously called natural killer Mycobacteria of the Study cell transcript 4, has recently been I Netea et al. explored the role of IL- The crucial role of IFN-γ in recognized as a proinfl ammatory 32 in the context of M. tuberculosis antimycobacterial immunity has cytokine (see Glossary) [1]. The main infection. In their study, freshly been shown in patients with defects sources of IL-32 are natural killer obtained human peripheral blood in the IFN-γ receptor [7]. Netea and cells, T cells, epithelial cells, and mononuclear cells (PBMCs) were colleagues’ fi ndings raise important blood monocytes. Four transcripts of stimulated with different TLR agonists, questions regarding (a) how IL-32 IL-32 are known at present. IL-32 has and gene expression and synthesis of levels vary in patients with defects in emerged as an important player in IL-32 was determined. The authors IFN-γ receptors who are susceptible innate and adaptive immune responses, showed that M. tuberculosis could elicit to tuberculosis and (b) how these and information is emerging on IL-32 release from PBMCs as well as variations may impact the course of synergism between IL-32 and other from purifed monocyte populations. the infection. One of the defi ciencies well-characterized players in innate of their study stems from the fact immunity. that the role of IL-32 in controlling The innate immune response M. -

O509 2-Hour Oral Session Frontiers in Tuberculosis the Role of Micrornas
O509 2-hour Oral Session Frontiers in tuberculosis The role of microRNAs in tuning the immune response in Mycobacterium tuberculosis infection Joyoti Basu*1 1Bose Institute, Chemistry, Kolkata, India Background: The role of post-transcriptional regulatory mechanisms in calibrating the immune response of macrophages infected with Mycobacterium tuberculosis (Mtb), remains poorly understood. We have performed global miRNA and transcriptome profiling to build an miRNA – mRNA network in Mtb- infected macrophages, and to understand its impact on the immune response to infection. Material/methods: We have performed global qRT-PCR profiling of miRNAs and microarray analysis of mRNAs to profile the transcriptome of macrophages infected with Mtb. Bioinformatic analyses has been performed using the tool MAGIA, to build an miRNA-mRNA network. We have then focused on selected miRNAs to analyze their effects on the immune response by analyzing pathways associated with inflammation and autophagy. Results: During Mtb infection, the let-7 miRNAs appeared as highly connected nodes of the miRNA- mRNA network. We establish that A 20 (TNFAIP3), a negative regulator of NF- B signaling is a target of let- 7. Augmented release of TNF-alfa, IL-1beta, CCL2, CXCL1, IL-6 and NO occurs in Mtb- infected macrophages in the absence of A20. During infection, downregulation of let -7f leads to concomitant rise in A20, likely tuning down inflammatory mediators. Transfection with a let -7f mimic, leads to increased cytokine release after infection. In addition to let-7f, miR-17 family members are also downregulated during Mtb infection of macrophages. We show that forced expression of miR-17 enhances autophagy during Mtb infection. -

Activity Report 2009 – 2010
Activity Report 2009 – 2010 L V Prasad Eye Institute Kallam Anji Reddy Campus L V Prasad Marg, Banjara Hills Hyderabad 500 034, India Tel: 91 40 3061 2345 Fax: 91 40 2354 8271 e-mail: [email protected] L V Prasad Eye Institute Patia, Bhubaneswar 751 024 Orissa, India Tel: 91 0674 3989 2020 Fax: 91 0674 3987 130 e-mail: [email protected] L V Prasad Eye Institute G M R Varalakshmi Campus Door No: 11-113/1 Hanumanthawaka Junction Visakhapatnam 530 040 Andhra Pradesh, India Tel: 91 0891 3989 2020 Fax: 91 0891 398 4444 L V Prasad Eye Institute e-mail: [email protected] Excellence • Equity • Effi ciency Art with vision, for vision Artist-in-residence Sisir Sahana in his workshop on A view of the Art Gallery on Level 6 at Hyderabad LVPEI’s Kallam Anji Reddy campus, Hyderabad creating campus, where several works by Mr Surya Prakash, one of his signature glass sculptures. Inset: A piece from our senior artist-in-residence are on display. his latest collection, entitled “The long climb”. Inset: The hand that wields the paintbrush! L V Prasad Eye Institute Committed to excellence and equity in eye care Activity Report April 2009 – March 2010 Collaborating Centre for Prevention of Blindness L V Prasad Eye Institute, a not-for-profi t charitable organization, is governed by two trusts: Hyderabad Eye Institute and Hyderabad Eye Research Foundation. Donations to Hyderabad Eye Research Foundation are 175% exempt under section 35 (i) (ii) and donations made to Hyderabad Eye Institute are 50% exempt under section 80G of the Income Tax Act. -

Year Book 2018 Year Book 2018
YEAR BOOK 2018 YEAR BOOK 2018 WEST BENGAL ACADEMY OF SCIENCE AND TECHNOLOGY CSIR-Indian Institute of Chemical Biology Jadavpur YEAR BOOK Kolkata 700 032 Registered under the West Bengal Act XXVI of 1961 (S/65001 of 1990-91) 2018 PAN – AAATW0707E Published by : Prof. Satyabrata Pal, Elected Member, ISI, FRSS Formerly, Dean, Post Graduate Studies, BCKV and Honorary Visiting Professor, ISI, Kolkata Editor, West Bengal Acadepmy of Science and Technology Assisted by : Dr. Arun Bandyopadhyay, Ph.D. Chief Scientist, CSIR-IICB, Kolkata-700 032 Secretary, West Bengal Academy of Science and Technology WAST Secretariat CSIR-Indian Institute of Chemical Biology 4, Raja S. C. Mullick Road WEST BENGAL Jadavpur, Kolkata 700 032 A C Telephone: (033) 2499-5796 A W A D e-mail: [email protected] E M Website: http://www.iicb.res.in/wast/index.html S T Y SCIENCE Printed by : WEST BENGAL ACADEMY OF SCIENCE AND TECHNOLOGY Creative Data Centre Registered Office : CSIR-Indian Institute of Chemical Biology 58/32, Prince Anwar Shah Road 4, Raja S. C. Mullick Road, Jadavpur Kolkata- 700 045 Kolkata 700 032 E-mail: [email protected] 1 2 YEAR BOOK 2018 YEAR BOOK 2018 AD-HOC Committee (1986-1989) Contents 1. Professor Sushil Kumar Mukherjee : Chairman 2. Professor Syama Pada Sen Introduction 5 3. Professor Asok Ghosh Memorandum of Association 6 4. Dr. Satyesh Chandra Pakrashi Rules and Regulations 9 Approved Amendments–I 25 5. Professor Subodh Kumar Roy Approved Amendments–II 29 6. Professor Asok Kumar Barua Past Office Bearers 34 7. Professor Nityananda Saha Council : 2016-2018 37 Sectional Committees : 2016-2018 39 8. -

Awardees of National Bioscience Award for Career Development
AWARDEES OF NATIONAL BIOSCIENCE AWARD FOR CAREER DEVELOPMENT Awardees for the year 2016 1. Dr. Mukesh Jain, Associate Professor, School of Computational and Integrative Sciences, Jawaharlal Nehru University, New Delhi-110067 2. Dr. Samir K. Maji, Associate Professor, Indian Institute of Technology, Powai, Mumbai- 400076 3. Dr. Anindita Ukil, Assistant Professor, Calcutta University, Kolkata 4. Dr. Arnab Mukhopadhyay, Staff Scientist V, National Institute of Immunology, Aruna Asaf Ali Marg, New Delhi- 110067 5. Dr. Rohit Srivastava, Professor, Indian Institute of Technology, Bombay, Mumbai- 400076 6. Dr. Pinaki Talukdar, Associate Professor, Indian Institute of Science Education and Research, Dr. Homi Bhabha Road, Pashan, Pune- 7. Dr. Rajnish Kumar Chaturvedi, Senior Scientist, CSIR- Indian Institute of Toxicology Research, Lucknow-226001 8. Dr. Jackson James, Scientist E-II, Neuro Stem Cell Biology Lab, Neurobiology Division, Rajiv Gandhi Centre for Biotechnology, Thiruvananthapuram, Kerala- 695014 Awardees for the year 2015 1. Dr. Sanjeev Das, Staff Scientist-V, National Institute of Immunology, New Delhi 2. Dr. Ganesh Nagaraju, Assistant Professor, Department of Biotechnology, Indian Institute of Science, Bangalore- 5600012. 3. Dr. Suvendra Nath Bhattacharya, Principal Scientist, CSIR- Indian Institute of Chemical Biology, Kolkata- 700032 4. Dr. Thulasiram H V, Principal Scientist, CSIR-National Chemical Laboratory, Pune- 411008. 5. Dr. Pawan Gupta, Principal Scientist, Institute of microbial Technology, Chandigarh- 160036. 6. Dr. Souvik Maiti, Principal Scientist, CSIR-Institute of Genomics and Integrative Biology, Delhi- 110025. 7. Dr. Pravindra Kumar, Associate Professor, Department of Biotechnology, IIT, Roorkee- 247667. 8. Dr. Anurag Agrawal, Principal Scientist, CSIR-Institute of Genomics and Integrative Biology, Delhi- 110025 9. Dr. Gridhar Kumar Pandey, Professor, Department of Plant Molecular Biology, University of Delhi South Campus, New Delhi- 110067 10. -

National Bioscience Awards for Career Development
AWARDEES OF NATIONAL BIOSCIENCE AWARDS FOR CAREER DEVELOPMENT Awardees for the year 2012 1. Dr. Kaustuv Sanyal, Associate Professor, Molecular Mycology Laboratory, Molecular Biology & Genetics Unit, Jawaharlal Nehru Centre for Advance Scientific Research, Jakkur P.O. Bangalore 560064 2. Dr Naval Kishore Vikram, Associate Professor, Department of Medicine, All India Institute of Medical Sciences (AIIMS), Ansari Nagar, New Delhi- 110029 3. Dr. Aditya Bhushan Pant, Senior Scientist & In-charge, In Vitro Toxicology Laboratory, Indian Institute of Toxicology Research, PO Box: 80, MG Marg, Lucknow 226001 (UP) India 4. Dr. Subrata Adak, Senior Scientist, Indian Institute of Chemical Biology; 4, Raja S.C. Mullick Road, Kolkata-700032 5. Dr. Durai Sundar, Assistant Professor, Dept of Biochemical Engineering & Biotechnology, Indian Institute of Technology (IIT) Delhi, Hauz Khas, New Delhi – 110016 6. Dr S Venkata Mohan, Senior Scientist, Bioengineering and Environmental Centre (BEEC) CSIR-Indian Institute of Chemical Technology, Hyderabad-500 607 7. Dr. Munia Ganguli, Scientist E-I, CSIR-Institute of Genomics & Integrative Biology, Mall Road,New Delhi 110 007 8. Dr. Asad U Khan, Associate Professor & Coordinator/Head of Biotechnology Department, A.M.U, Interdisciplinary Biotechnology Unit, A.M.U., Aligarh 202002 9. Dr. Sathees C. Raghavan, Assistant Professor, Department of Biochemistry, Indian Institute of Science, Bangalore 560 012 10. Dr. Vidita A. Vaidya, Associate Professor, Department of Biological Sciences, Tata Institute of Fundamental Research, 1, Homi Bhabha Road, Colaba, Mumbai - 400005 Awardees for the year 2011 1. Dr. M. M. Parida, Scientist-F, Joint Director, Division of Virology Defence R & D Establishment, DRDE, DRDO, Ministry of Defence, Jhansi Road, Gwalior- 474002 2. -

Book Download
SOCIETY OF BIOLOGICAL CHEMISTS (INDIA) (1930 – 2011) 1 TABLE OF CONTENTS 1. Goals and activities of SBC(I) 2. Rules and Bye-laws of SBC(I) 3. Past Presidents, Secretaries, Treasurers (with tenure) 4. “Reminiscences on the development of the Society of Biological Chemists (India): a personal perspective” by Prof. N. Appaji Rao 5. “Growth of Biochemistry in India” by Prof. G. Padmanaban 6. Current office bearers 7. Current Executive Committee Members 8. Office staff 9. Past meeting venues of SBC(I) 10. SBC(I) awards, criteria and procedure for applying 11. SBC(I) awardees 12. Current list of life members with address 13. Acknowledgments 2 GOALS AND ACTIVITIES OF SBC(I) To meet a long felt need of scientists working in the discipline of biological chemistry " The Society Of Biological Chemists (India)" was founded in 1930, with its Head Quarters at Indian Institute of Science, Bangalore. It was registered under the Societies Act in the then princely state of Mysore and the memorandum of registration was signed by the late Profs. V. Subramanian, V. N. Patwardhan and C. V. Natarajan, who were leading personalities in the scientific firmament during that period. The Society played a crucial role during the Second World War by advising the Government on the utilization of indigenous biomaterials as food substitutes, drugs and tonics, on the industrial and agricultural waste utilization and on management of water resources. The other areas of vital interest to the Society in the early years were nutrition, proteins, enzymes, applied microbiology, preventive medicines and the development of high quality proteins from indigenous plant sources.