Introduction to Python
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Python Qt Tutorial Documentation Release 0.0
Python Qt tutorial Documentation Release 0.0 Thomas P. Robitaille Jun 11, 2018 Contents 1 Installing 3 2 Part 1 - Hello, World! 5 3 Part 2 - Buttons and events 7 4 Part 3 - Laying out widgets 9 5 Part 4 - Dynamically updating widgets 13 i ii Python Qt tutorial Documentation, Release 0.0 This is a short tutorial on using Qt from Python. There are two main versions of Qt in use (Qt4 and Qt5) and several Python libraries to use Qt from Python (PyQt and PySide), but rather than picking one of these, this tutorial makes use of the QtPy package which provides a way to use whatever Python Qt package is available. This is not meant to be a completely exhaustive tutorial but just a place to start if you’ve never done Qt development before, and it will be expanded over time. Contents 1 Python Qt tutorial Documentation, Release 0.0 2 Contents CHAPTER 1 Installing 1.1 conda If you use conda to manage your Python environment (for example as part of Anaconda or Miniconda), you can easily install Qt, PyQt5, and QtPy (a common interface to all Python Qt bindings) using: conda install pyqt qtpy 1.2 pip If you don’t make use of conda, an easy way to install Qt, PyQt5, and QtPy is to do: pip install pyqt5 qtpy 3 Python Qt tutorial Documentation, Release 0.0 4 Chapter 1. Installing CHAPTER 2 Part 1 - Hello, World! To start off, we need to create a QApplication object, which represents the overall application: from qtpy.QtWidgets import QApplication app= QApplication([]) You will always need to ensure that a QApplication object exists, otherwise your Python script will terminate with an error if you try and use any other Qt objects. -

Testing Pyside/Pyqt Code Using the Pytest Framework and Pytest-Qt
Testing PySide/PyQt Code Using the pytest framework and pytest-qt Florian Bruhin “The Compiler” Bruhin Software 06. November 2019 Qt World Summit, Berlin About me • 2011: Started using Python • 2013: Started using PyQt and developing qutebrowser • 2015: Switched to pytest, ended up as a maintainer • 2017: qutebrowser v1.0.0, QtWebEngine by default • 2019: 40% employed, 60% open-source and freelancing (Bruhin Software) Giving trainings and talks at various conferences and companies! Relevant Python features Decorators registered_functions: List[Callable] = [] def register(f: Callable) -> Callable: registered_functions.append(f) return f @register def func() -> None: .... Relevant Python features Context Managers def write_file() -> None: with open("test.txt", "w") as f: f.write("Hello World") Defining your own: Object with special __enter__ and __exit__ methods. Relevant Python features Generators/yield def gen_values() -> Iterable[int] for i in range(4): yield i print(gen_values()) # <generator object gen_values at 0x...> print(list(gen_values())) # [0, 1, 2, 3] PyQt • Started in 1998 (!) by Riverbank Computing • GPL/commercial • Qt4 $ PyQt4 Qt5 $ PyQt5 PySide / Qt for Python • Started in 2009 by Nokia • Unmaintained for a long time • Since 2016: Officially maintained by the Qt Company again • LGPL/commercial • Qt4 $ PySide Qt5 $ PySide2 (Qt for Python) Qt and Python import sys from PySide2.QtWidgets import QApplication, QWidget, QPushButton if __name__ == "__main__": app = QApplication(sys.argv) window = QWidget() button = QPushButton("Don't -

Asyncprotect T1 Extunderscore Gui Documentation
async_gui Documentation Release 0.1.1 Roman Haritonov Aug 21, 2017 Contents 1 Usage 1 1.1 Installation................................................1 1.2 First steps.................................................1 1.3 Tasks in threads.............................................2 1.4 Tasks in processes............................................3 1.5 Tasks in greenlets.............................................3 1.6 Returning result.............................................3 2 Supported GUI toolkits 5 3 API 7 3.1 engine..................................................7 3.2 tasks...................................................8 3.3 gevent_tasks...............................................9 4 Indices and tables 11 Python Module Index 13 i ii CHAPTER 1 Usage Installation Install with pip or easy_install: $ pip install --upgrade async_gui or download latest version from GitHub: $ git clone https://github.com/reclosedev/async_gui.git $ cd async_gui $ python setup.py install To run tests: $ python setup.py test First steps First create Engine instance corresponding to your GUI toolkit (see Supported GUI toolkits): from async_gui.tasks import Task from async_gui.toolkits.pyqt import PyQtEngine engine= PyQtEngine() It contains decorator @engine.async which allows you to write asynchronous code in serial way without callbacks. Example button click handler: @engine.async def on_button_click(self, *args): self.status_label.setText("Downloading image...") 1 async_gui Documentation, Release 0.1.1 # Run single task in separate thread -

CIS192 Python Programming Graphical User Interfaces
CIS192 Python Programming Graphical User Interfaces Robert Rand University of Pennsylvania November 19, 2015 Robert Rand (University of Pennsylvania) CIS 192 November 19, 2015 1 / 20 Outline 1 Graphical User Interface Tkinter Other Graphics Modules 2 Text User Interface curses Other Text Interface Modules Robert Rand (University of Pennsylvania) CIS 192 November 19, 2015 2 / 20 Tkinter The module Tkinter is a wrapper of the graphics library Tcl/Tk Why choose Tkinter over other graphics modules I It’s bundled with Python so you don’t need to install anything I It’s fast I Guido van Rossum helped write the Python interface The docs for Tkinter aren’t that good I The docs for Tk/Tcl are much better I Tk/Tcl functions translate well to Tkinter I It’s helpful to learn the basic syntax of Tk/Tcl Tk/Tcl syntax ! Python: I class .var_name -key1 val1 -key2 val2 ! var_name = class(key1=val1, key2=val2) I .var_name method -key val ! var_name.method(key=val) Robert Rand (University of Pennsylvania) CIS 192 November 19, 2015 3 / 20 Bare Bones Tkinter from Tkinter import Frame class SomeApp(Frame): def __init__(self, master=None): tk.Frame.__init__(self, master) def main(): root = tk.Tk() app = SomeApp(master=root) app.mainloop() if __name__ ==’__main__’: main() Robert Rand (University of Pennsylvania) CIS 192 November 19, 2015 4 / 20 Images By default, Tkinter only supports bitmap, gif, and ppm/pgm images More images are supported with Pillow Pillow is a fork of Python Imaging Library pip install pillow from PIL import Image, ImageTk Create a PIL -

Multiplatformní GUI Toolkity GTK+ a Qt
Multiplatformní GUI toolkity GTK+ a Qt Jan Outrata KATEDRA INFORMATIKY UNIVERZITA PALACKÉHO V OLOMOUCI GUI toolkit (widget toolkit) (1) = programová knihovna (nebo kolekce knihoven) implementující prvky GUI = widgety (tlačítka, seznamy, menu, posuvník, bary, dialog, okno atd.) a umožňující tvorbu GUI (grafického uživatelského rozhraní) aplikace vlastní jednotný nebo nativní (pro platformu/systém) vzhled widgetů, možnost stylování nízkoúrovňové (Xt a Xlib v X Windows System a libwayland ve Waylandu na unixových systémech, GDI Windows API, Quartz a Carbon v Apple Mac OS) a vysokoúrovňové (MFC, WTL, WPF a Windows Forms v MS Windows, Cocoa v Apple Mac OS X, Motif/Lesstif, Xaw a XForms na unixových systémech) multiplatformní = pro více platforem (MS Windows, GNU/Linux, Apple Mac OS X, mobilní) nebo platformově nezávislé (Java) – aplikace může být také (většinou) událostmi řízené programování (event-driven programming) – toolkit v hlavní smyčce zachytává události (uživatelské od myši nebo klávesnice, od časovače, systému, aplikace samotné atd.) a umožňuje implementaci vlastních obsluh (even handler, callback function), objektově orientované programování (objekty = widgety aj.) – nevyžaduje OO programovací jazyk! Jan Outrata (Univerzita Palackého v Olomouci) Multiplatformní GUI toolkity duben 2015 1 / 10 GUI toolkit (widget toolkit) (2) language binding = API (aplikační programové rozhraní) toolkitu v jiném prog. jazyce než původní API a toolkit samotný GUI designer/builder = WYSIWYG nástroj pro tvorbu GUI s využitím toolkitu, hierarchicky skládáním prvků, z uloženého XML pak generuje kód nebo GUI vytvoří za běhu aplikace nekomerční (GNU (L)GPL, MIT, open source) i komerční licence např. GTK+ (C), Qt (C++), wxWidgets (C++), FLTK (C++), CEGUI (C++), Swing/JFC (Java), SWT (Java), JavaFX (Java), Tcl/Tk (Tcl), XUL (XML) aj. -

Writing Standalone Qt & Python Applications for Android
Writing standalone Qt & Python applications for Android Martin Kolman Red Hat & Faculty of Informatics, Masaryk University http://www.modrana.org/om2013 [email protected] 1 Overview Writing Android applications with Python and Qt How it works The pydroid project Examples Acknowledgement 2 Writing Android applications with Python and Qt Lets put it all together ! So that: applications can be written entirely in Python Qt/QML can be used for the GUI the result is a standalone Google Play compatible APK package deployment is as easy as possible all binary components can be recompiled at will 3 Writing Android applications with Python and Qt Writing Android applications with Python and Qt Python has been available for Android for ages the Qt graphics library has been available for Android since early 2011 there have been some proof of concepts of Qt-Python bindings working on Android and even some proof of concepts of distributing Python + Qt applications 4 How it works How “normal” Android applications look like written in the Android Java running on the Dalvik VM can have a native backend written in C/C++ using the Android NDK application packages are distributed as files with the .apk extension there is no support for cross-package dependencies 5 How it works Necessitas - Qt for Android provides Qt libraries compiled for Android 2.2+ most of Qt functionality is supported, including OpenGL acceleration and Qt Quick handles automatic library downloads & updates through the Ministro service 6 How it works Python -

Pyside Overview
PySide overview Marc Poinot (ONERA/DSNA) Outline Quite short but practical overview ▶Qt ■ Toolkit overview ■ Model/View ▶PySide ■ pyQt4 vs PySide ■ Designer & Cython ■ Widget bindings ■ Class reuse PySide - Introduction ONERA/PySide-2/8 [email protected] Qt - Facts ▶Qt is cute ■ Cross platform application framework for GUI X Window System, Windows... ■ C++ kernel + many bindings Including Python ■ Release v5.3 05/2014 ■ License GPL+LGPL+Commercial Contact your own lawyer... ▶Components ■ Core: QtCore, QtGui... ■ Specialized: QtWebKit, QtSVG, QtSQL... ■ Tools : Creator, Designer... PySide - Introduction ONERA/PySide-3/8 [email protected] Qt - Example PySide - Introduction ONERA/PySide-4/8 [email protected] Python bindings ▶pyQt ■ The first to come Some services have hard coded import PyQt4 ■ GPL - Use only in free software ▶PySide ■ Some syntactic & behavior differences ■ LGPL - Use allowed in proprietary software PySide overview hereafter mixes Qt/PySide features PySide - Introduction ONERA/PySide-5/8 [email protected] PySide - Facts ▶Full Python binding ■ Qt classes as Python classes ■ Python types as parameter types ▶Release 1.2.2 04/2014 ▶Reference documentation http://pyside.github.io/docs/pyside/ ▶Production process ■ Uses many steps ■ Better with setup & source management PySide - Introduction ONERA/PySide-6/8 [email protected] PySide - Production process W class Ui_W class WB(QWidget,Ui_W) ☺ A.ui uic ui_A.pyx Designer B.py cython A.c A.so A.rc rcc Res_rc.py PySide - Introduction ONERA/PySide-7/8 [email protected] -

Pyqt-Käyttöliittymäkirjasto Pyqt-Käyttöliittymäkirjasto
PyQt-käyttöliittymäkirjasto PyQt-käyttöliittymäkirjasto • PyQt on laaja ja yleinen kirjasto – Tehokas grafiikka ja monipuoliset ominaisuudet – Toimii monilla eri alustoilla – PyQt5 pitää sisällään 620 luokkaa ja 6000 funktiota ja metodia • Muita vastaavia käyttöliittymäkirjastoja: – PySide, PyGTK, wxPython, Tkinter • Qt on käytettävissä monesta muustakin kielestä – PyQt on joukko sidontoja alla olevaan Qt-toteutukseen – PyQt on toteutettu joukkona Python moduuleita • Keskeinen käsite ovat käyttöliittymäkomponentit (widget) – Ne vastaavat usein suoraan ikkunaan ilmestyviä graafisia olioita 2 Esimerkki: proseduaalinen ohjelmointi • Proseduraalisessa ohjelmoinnissa luodaan aliohjelmia – Tyypillisesti muuttavat ohjelman tietorakenteiden tilaa • Oheinen luo ikkunan PyQt5:llä import sys from PyQt5.QtWidgets import QWidget, QPushButton, QApplication def Example(): app = QApplication(sys.argv) w = QWidget() b = QPushButton('Push', w) b.clicked.connect(w.close) w.show() sys.exit(app.exec_()) 3 Esimerkki: sama olio-ohjelmoinnilla • Olio-ohjelmoinnissa luomme luokan – Perustoiminnallisuus rakennetaan olioon, jota käytetään • Ohessa widget-olion kutsuminen yms. on jätetty pois class Example(QWidget): def __init__(self): super().__init__() self.initUI() def initUI(self): self.__btn = QPushButton('Push', self) self.show() self.activate_close() def activate_close(self): self.__btn.clicked.connect(self.close) self.show() 4 Havaintoja edellisten esimerkkien eroista • Proseduraaliversiossa voisimme antaa argumentteja – Näin aliohjelma olisi yleiskäyttöisempi -

Qte-Doc Documentation Release 0.5.0
qte-doc Documentation Release 0.5.0 David Townshend September 06, 2012 Contents 1 Key features 3 2 Installation 5 3 Table of contents 7 3.1 Tutorial................................................7 3.2 UIs and Resources..........................................8 3.3 Edit Widgets............................................. 11 3.4 Model/View framework....................................... 14 3.5 Core Tools.............................................. 21 Python Module Index 25 i ii qte-doc Documentation, Release 0.5.0 qte is a wrapper around the PySide GUI toolkit making it simpler and more pythonic to use. No changes are made to the original PySide classes, but new classes and functions are introduced which effectively replace much of the most commonly used PySide GUI functionality. Contents 1 qte-doc Documentation, Release 0.5.0 2 Contents CHAPTER 1 Key features • All objects (i.e. those contained in PySide.QtGui, PySide.QtCore, and the new objects provided by this package) can be imported from a single namespace, e.g: >>> from qte import QRect, QWidget, DataModel Or, more commonly: >>> import qte >>> rect= qte.QRect() The PySide objects, i.e. those prefixed with “Q”, are exactly the same as those imported directly from PySide, all changes are in the new classes. • A number of convenience function had been added. See Core Tools for details. • The PySide model/view framework has been extended and simplified. See Model/View framework for more information. • A new singleton Application class is available, extending the functionality of QApplication. See Application and runApp for more information. •A Document class is provided to manage typical document application functions, such as opening, saving, etc. 3 qte-doc Documentation, Release 0.5.0 4 Chapter 1. -

Rapid GUI Application Development with Python
Rapid GUI Application Development with Python Volker Kuhlmann Kiwi PyCon 2009 — Christchurch 7 November 2009 Copyright c 2009 by Volker Kuhlmann Released under Creative Commons Attribution Non-commercial Share-alike 3.0[1] Abstract Options and tools for rapid desktop GUI application development using Python are examined, with a list of options for each stage of the tool chain. Keywords kiwipycon, kiwipycon09, Python, GUI, RAD, GUI toolkit, GUI builder, IDE, application development, multi-platform 1 Introduction GUI options are examined for Python with respect to performance, ease of use, runtime, and platform portabil- ity. Emphasis is put on platform-independence and free open source, however, commercial options are included. This paper is a summary of the conference presentation. The related slides are available online[2]. A full paper is planned. The following components of the tool chain are discussed: • Python interpreter • GUI toolkit • IDE (Integrated Development Environment) • GUI builder • Debugger • Database • Testing • Application packaging 2 Python Interpreter Getting the Python interpreter up and running is the easiest part of the tool chain, and is achieved by simply in- stalling the latest stable version for your platform—currently 2.6 or 3.1. The 3.x version is not totally backwards compatible. Pre-compiled packages are available for each platform. For a well-written easy to understand introduction to Python programming, see the book by Hetland[3]. 1 3 GUI toolkit The GUI toolkit provides the graphical user interface (GUI) elements, or widgets, like buttons, scrollbars, text boxes, label fields, tree view lists, file-open dialogues and so on. These widgets are an integral and essential part of the user interface. -

Python Programming — GUI
Python programming | GUI Finn Arup˚ Nielsen Department of Informatics and Mathematical Modelling Technical University of Denmark October 1, 2012 Python programming | GUI Graphical user interface Tk Python module Tkinter. Quick and dirty. (Langtangen, 2005, Chapters 6 & 11) wxWidgets (wxWindows) wxPython (Ubuntu package python-wxtools) and PythonCard (Ubuntu meta- package pythoncard) Others PyGTK for Gtk, PyQT and PySide for Qt Finn Arup˚ Nielsen 1 October 1, 2012 Python programming | GUI Tkinter Hello world from Tkinter import * def hello(): """Callback function for button press.""" print "Hello World" # Main root window root = Tk() # A button with a callback function Button(root, text="Press here", command=hello).pack() # .pack() method is necessary to make the button appear # Event loop root.mainloop() Finn Arup˚ Nielsen 2 October 1, 2012 Python programming | GUI Tkinter Hello world Finn Arup˚ Nielsen 3 October 1, 2012 Python programming | GUI Tkinter: menu, label Build a GUI for showing one tweet stored in a Mongo database. One tweets at a time. from Tkinter import * import pymongo, re pattern = re.compile(r'\bRT\b', re.UNICODE) Should read from the Mongo database, and display some of the fields in a graphical user interface. With a button the user should be able to click to the next item. Finn Arup˚ Nielsen 4 October 1, 2012 Python programming | GUI A callback function counter = 0 def next(event=None): """Function that can be used as a callback function from a button.""" global counter # Global variable not so pretty counter += 1 tweet -
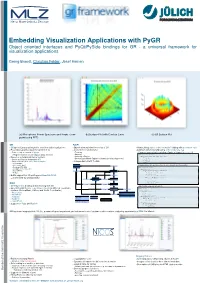
Object Oriented Interfaces and Pyqt/Pyside Bindings for GR
Embedding Visualization Applications with PyGR Object oriented interfaces and PyQt/PySide bindings for GR - a universal framework for visualization applications Georg Brandl, Christian Felder, Josef Heinen (a) Microphone Power Spectrum and Peaks (com- (b)Surface Plot with Contour Lines (c)3D Surface Plot puted using FFT) GR PyGR GRaphics Library optimized for real-time data visualizations Object oriented interface on top of GR Manipulating object states instead of fiddling with procedure calls • • • Procedural graphics backend (written in C) Convenience functions for Adding new functionality using derived objects, e.g. • • • Does not rely on creation of figures Zooming Clamp minimum values for selecting a Region of Interest (x , y > 0) • • • min min Panning 1 class ContourAxes(PlotAxes): Representation of continuous data streams • ) Selecting a Region def setWindow(self, xmin, xmax, ymin, ymax): Based on a Graphical Kernel System • if xmin < 1: • Detecting predefined Regions Of Interest (point in polygon test) xmin = 1 Device and Platform independent API • 6 • if ymin < 1: Large scale of logical device drivers/plugins: Independent of GUI Toolkits ymin = 1 • • return PlotAxes.setWindow(self, xmin, xmax, ymin, ymax) Qt, wxWidgets • X11, Quartz, Win32 Drawing a bar at a given position xi, yi to indicate a peak position • • PostScript, PDF, SVG class PeakBars(PlotCurve): • 2 GS (BMP, JPEG, PNG, TIFF) • def drawGR(self): MOV (MPEG4) if self.linetype is not None and self.x: • # preserve old values HTML5 lcolor = gr.inqlinecolorind() • 7 gr.setlinewidth(2) Builtin support for 2D plotting and OpenGL (GR3) gr.setlinecolorind(self.linecolor) • for xi, yi in zip(self.x, self.y): Coexistent 2D and 3D world gr.polyline([xi, xi], [0, yi]) ) 12 # restore old values gr.setlinecolorind(lcolor) QtGR gr.setlinewidth(1) Qt Widgets for drawing and interacting with GR Hide/Unhide dependent objects • • Specialized Qt Events, e.